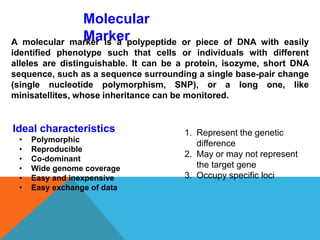
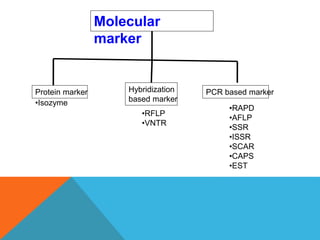
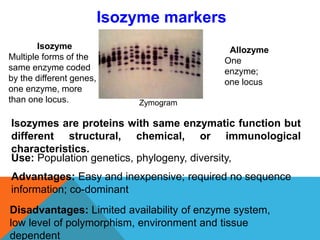
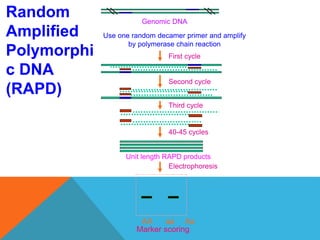
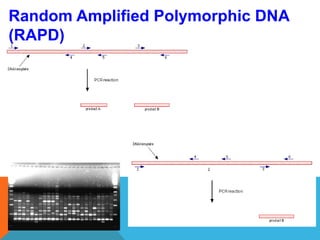
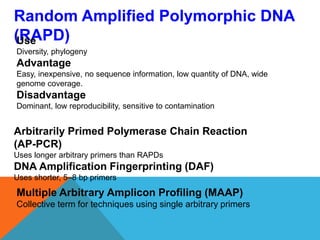
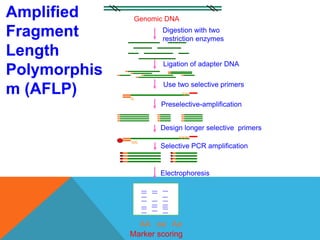
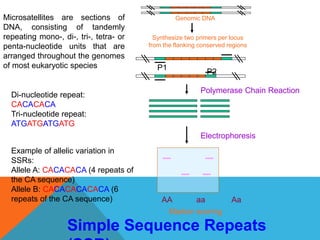
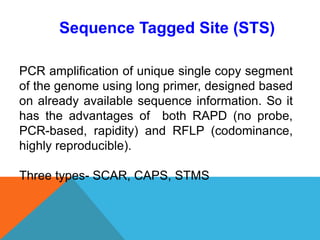
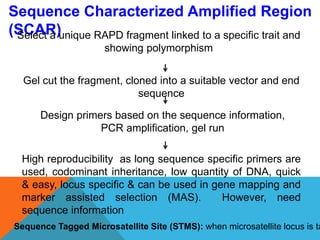
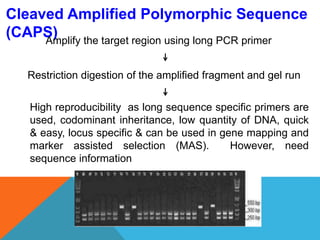
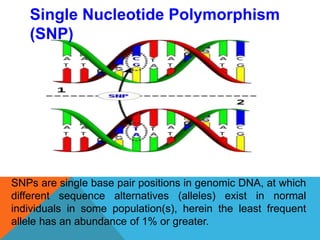
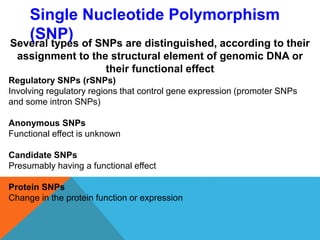
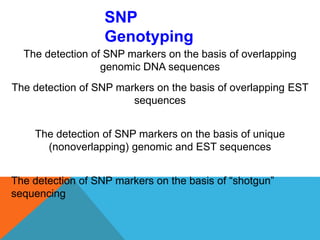
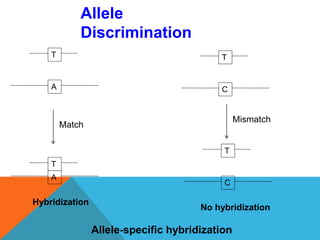
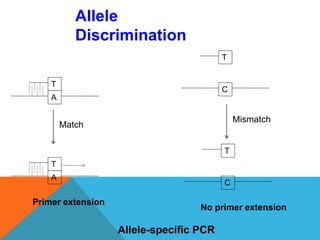
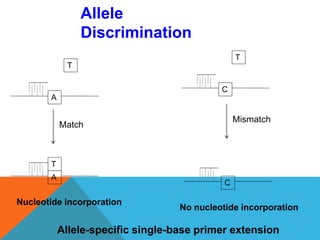
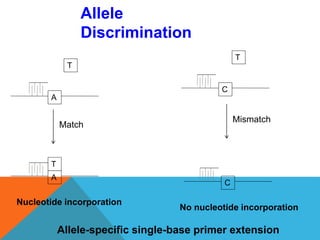
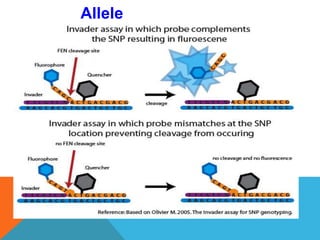
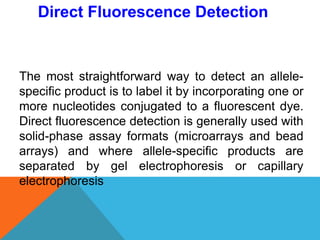
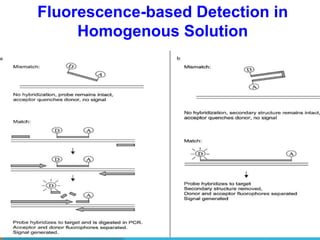
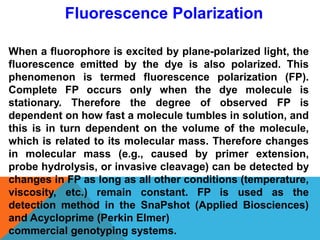
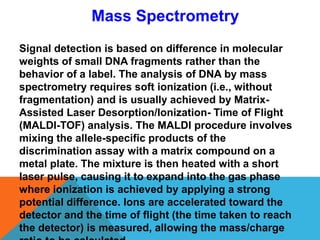
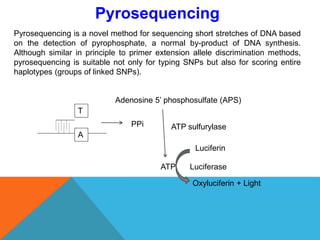
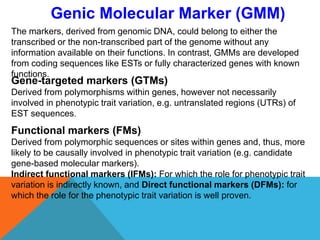
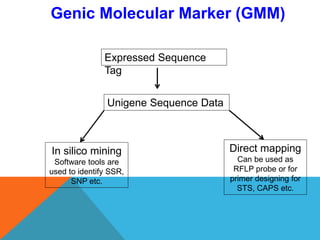
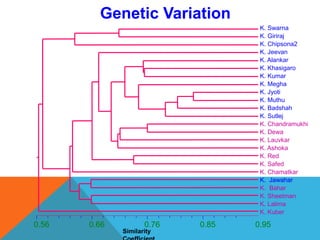
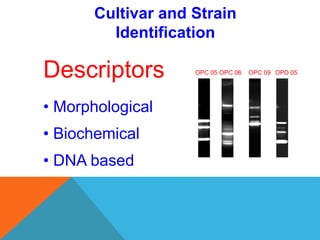
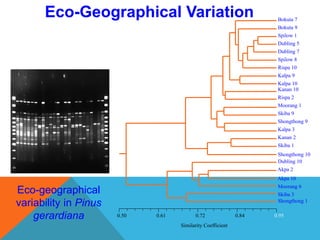
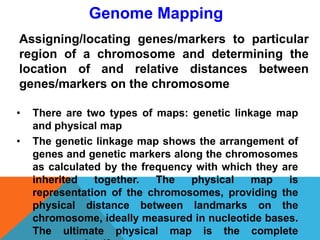
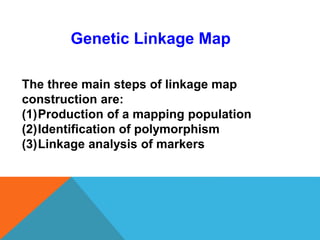
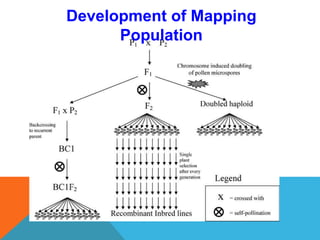
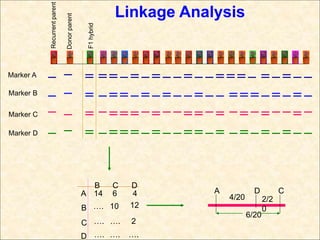
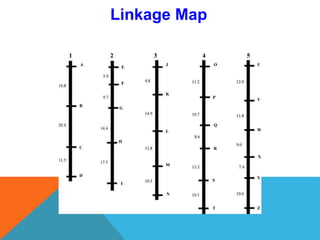
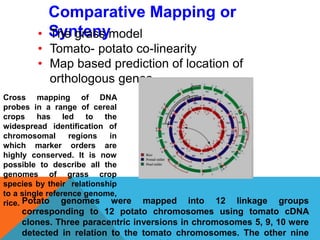
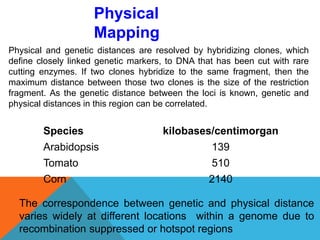
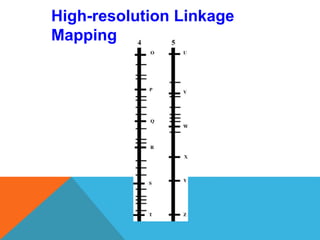
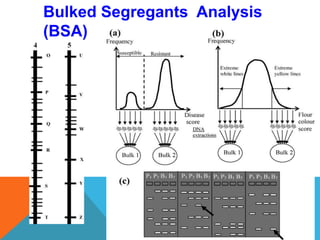
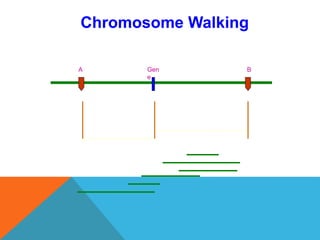
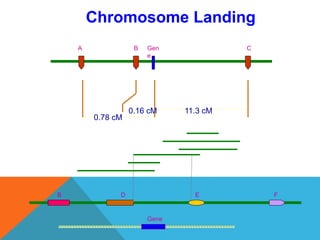
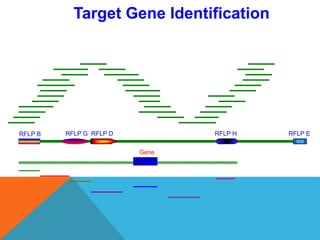
1. Molecular markers are DNA sequences that can be used to identify specific locations or genes on chromosomes. Different types of molecular markers include isozymes, restriction fragment length polymorphisms (RFLPs), variable number of tandem repeats (VNTRs), and single nucleotide polymorphisms (SNPs).
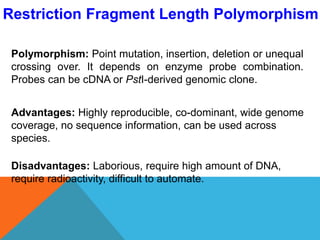
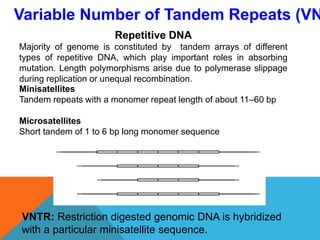
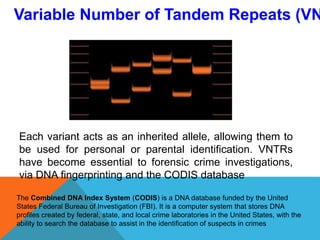
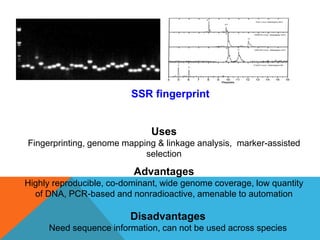
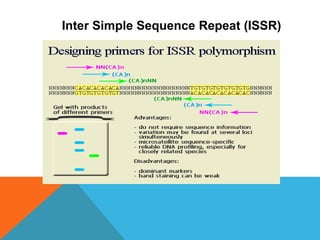
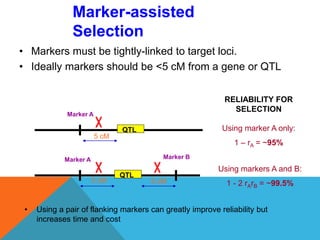
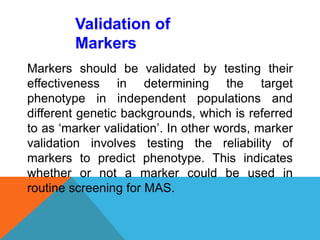
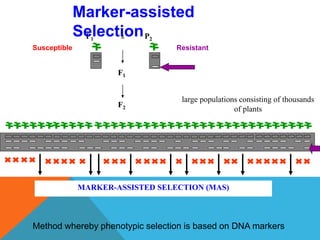
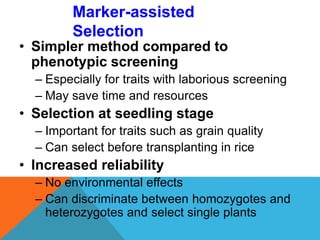
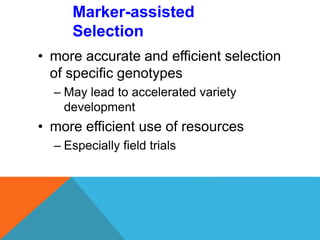
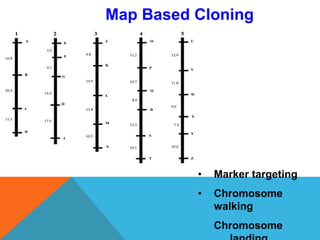
2. Molecular markers have a variety of uses in genetics and forensic analysis. RFLPs and VNTRs can be used for DNA fingerprinting and identifying individuals. SNP markers allow for analysis of genetic variations between individuals and populations. Molecular markers are also used for genome mapping and marker-assisted breeding in plants and animals.
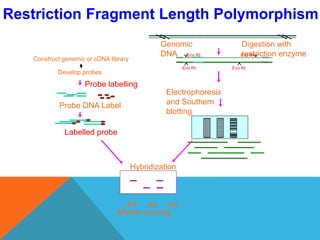
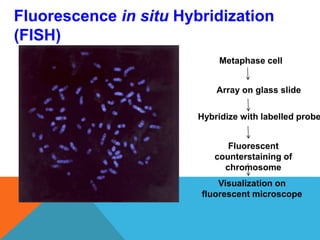
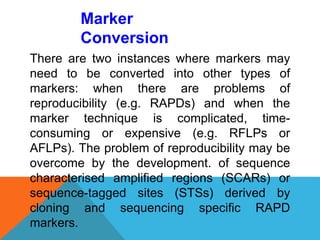
3. There are several techniques used to detect molecular markers, including gel electrophoresis, Southern blot