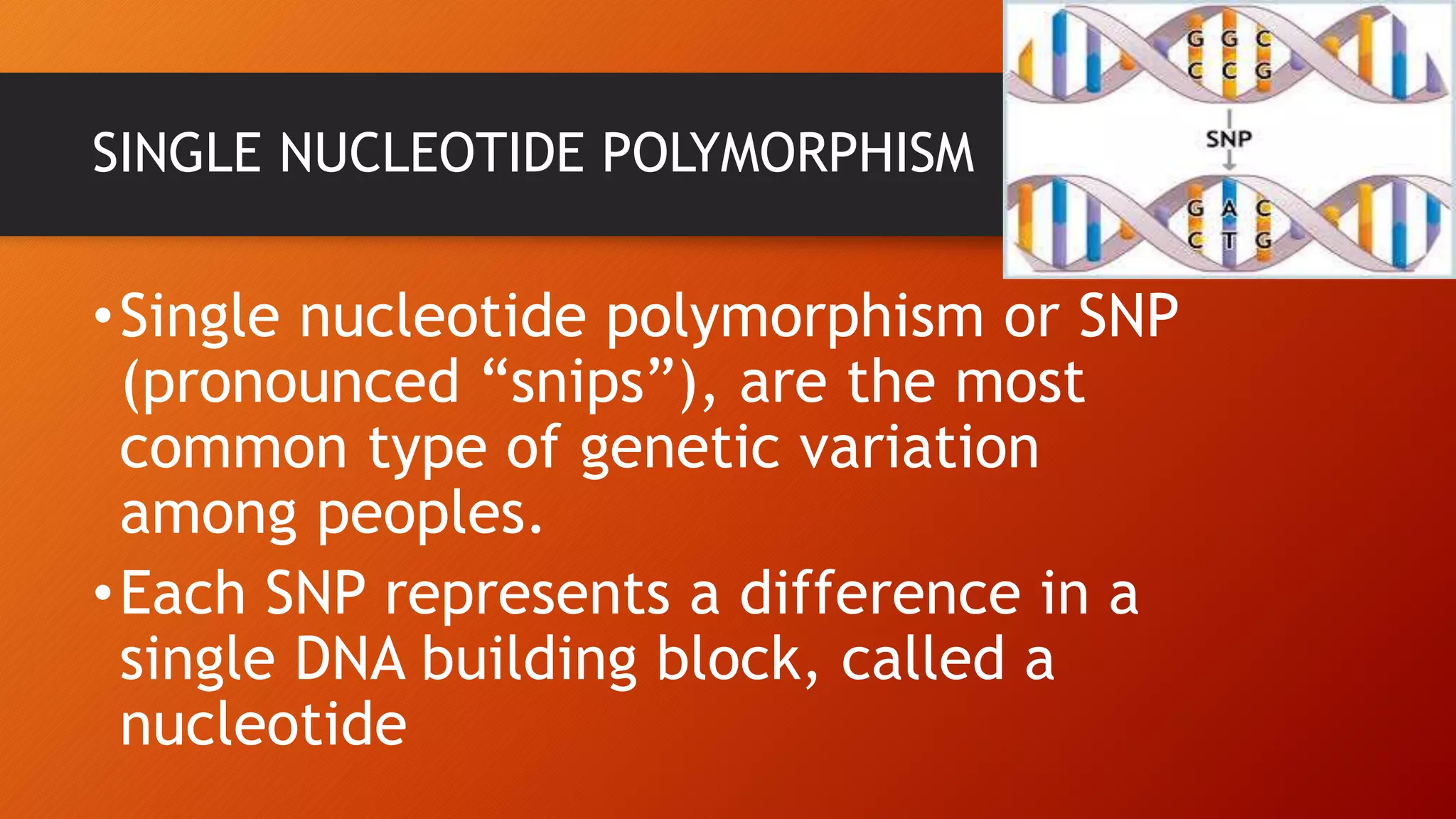
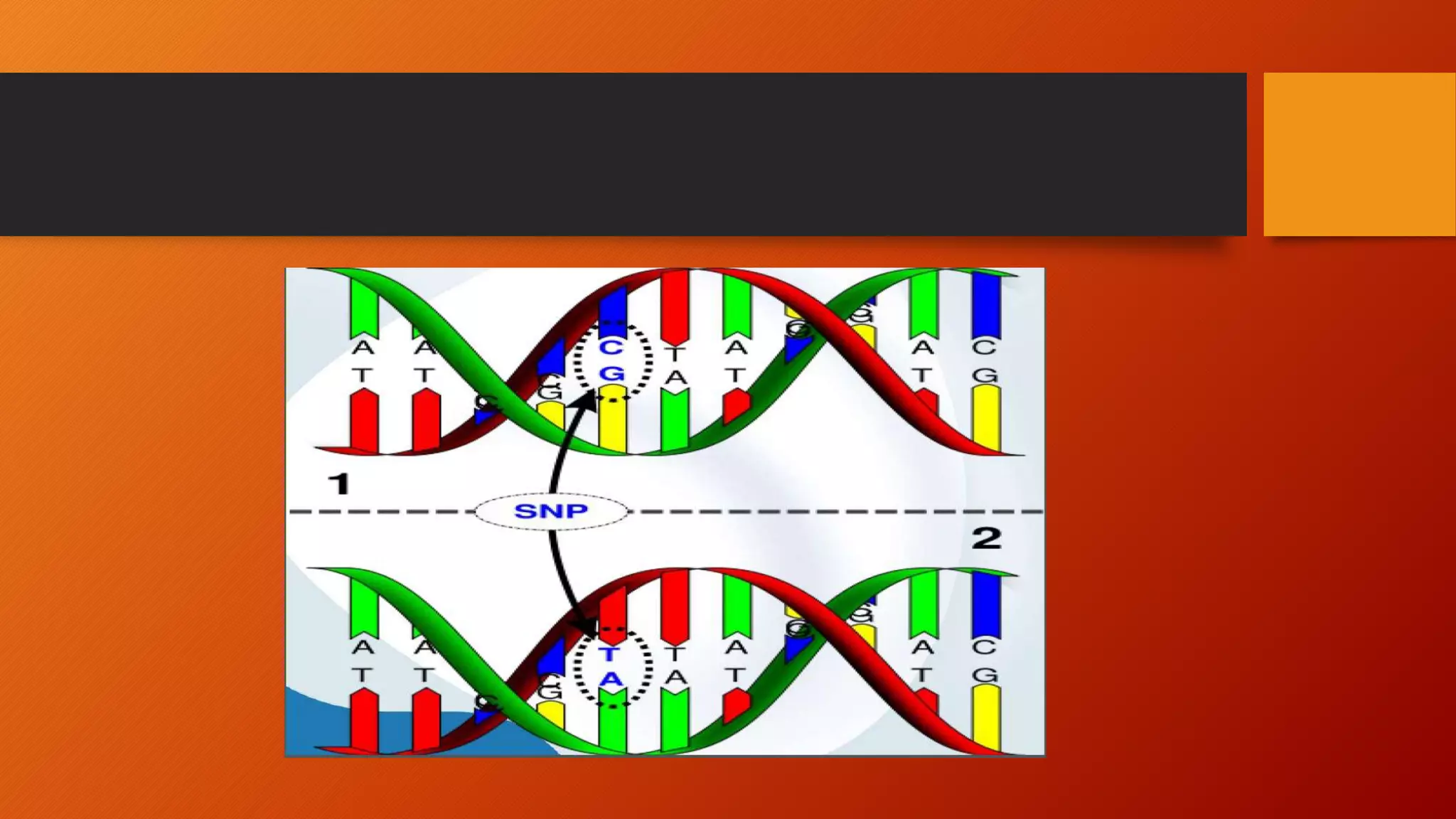
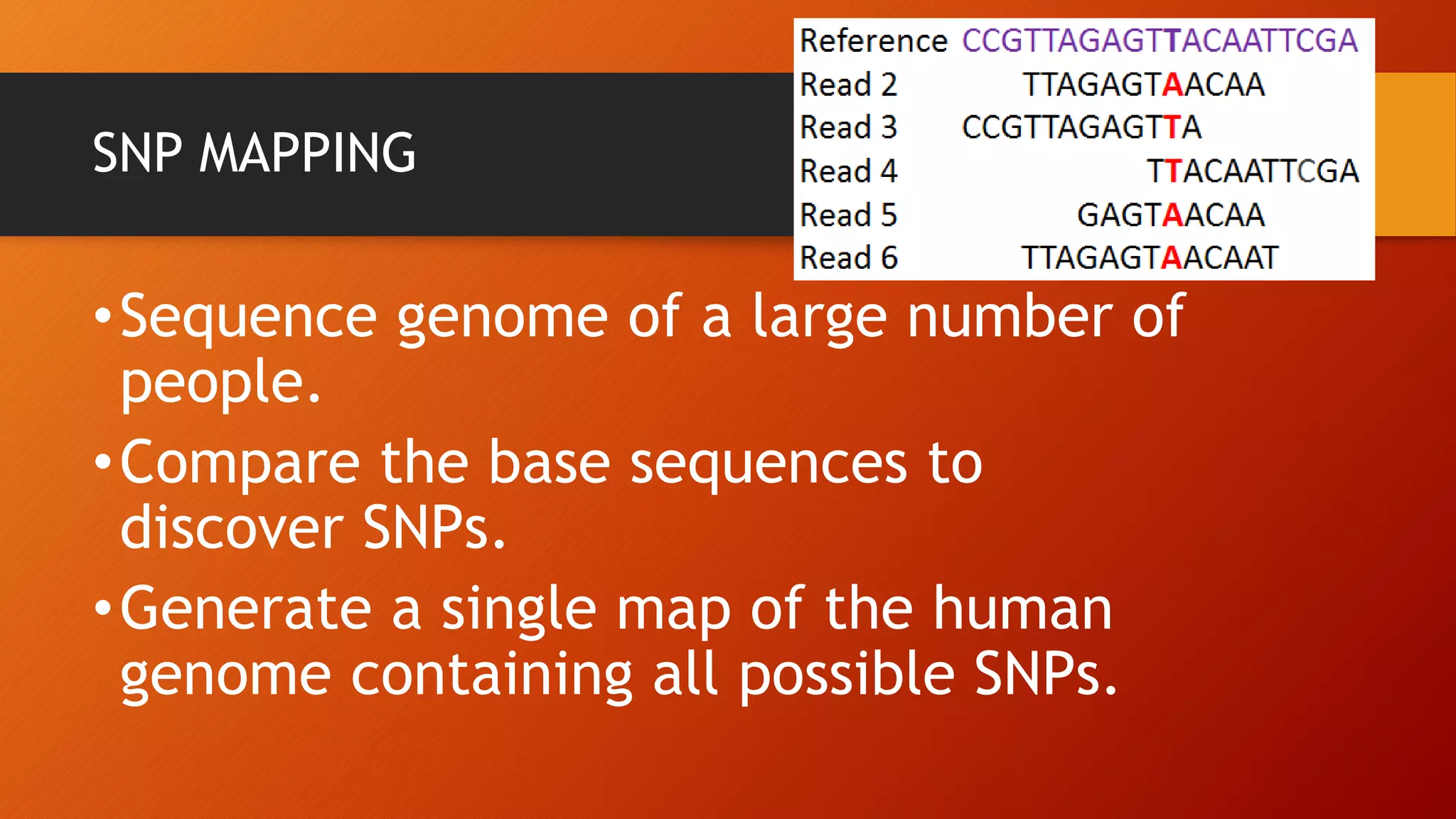
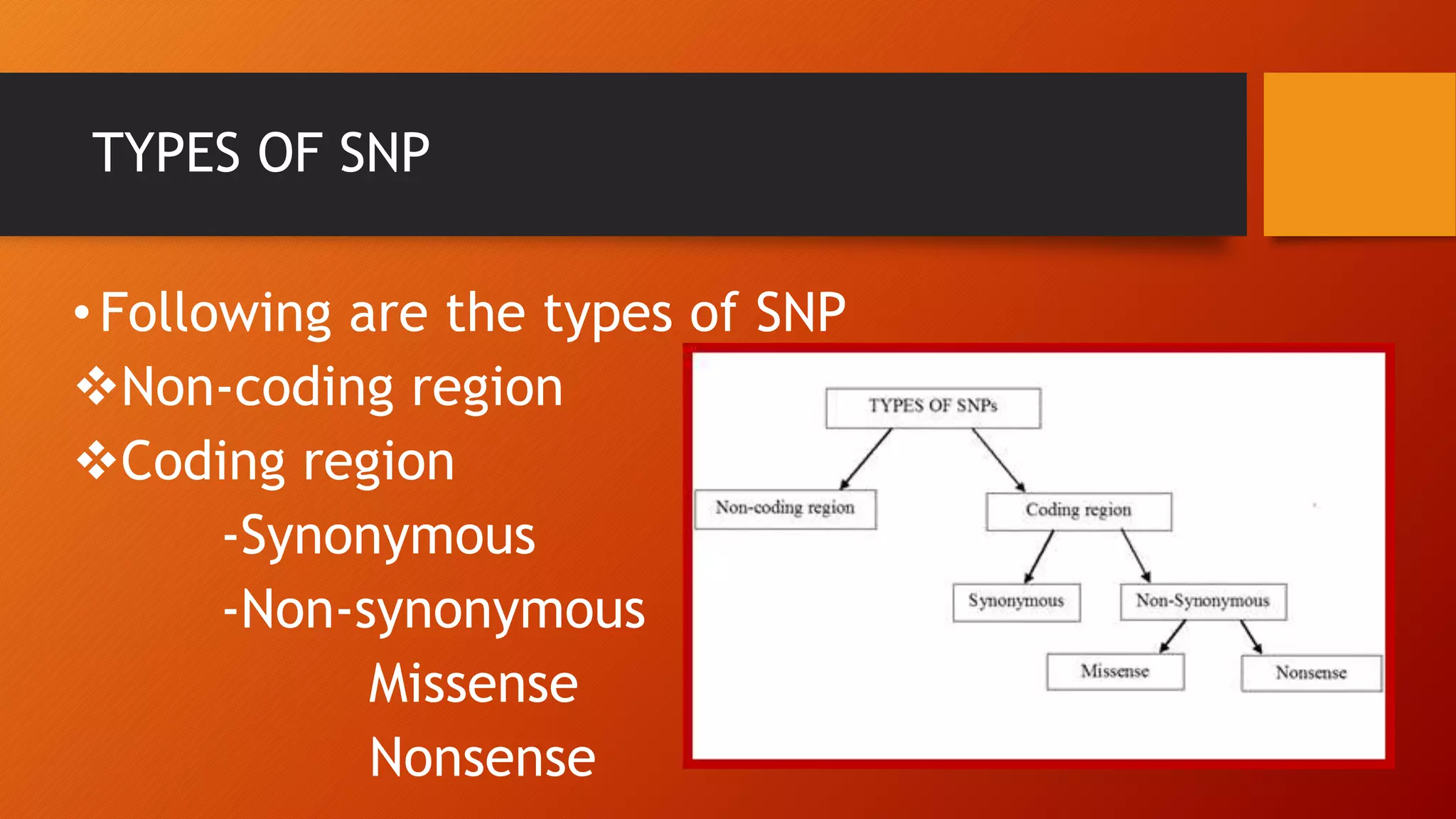
This document discusses single nucleotide polymorphisms (SNPs), which are variations in DNA sequences among individuals. SNPs are the most common type of genetic variation, occurring around every 100 to 300 bases. They can be found in coding and non-coding regions, and may alter protein function directly by changing amino acids or indirectly by affecting regulatory sequences. SNPs can influence disease susceptibility and treatment response. Methods for identifying SNPs involve detecting known SNPs using techniques like restriction enzyme analysis or sequencing to find new SNPs. SNPs have many applications including gene mapping, disease association studies, diagnostics, and predicting treatment response.