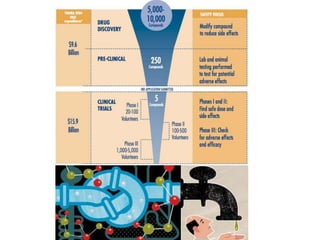
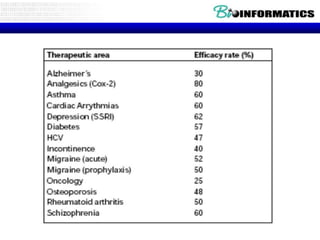
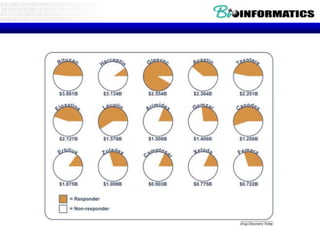
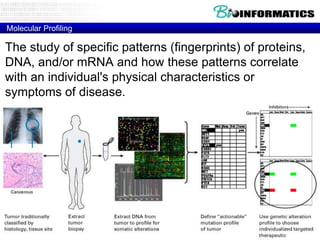
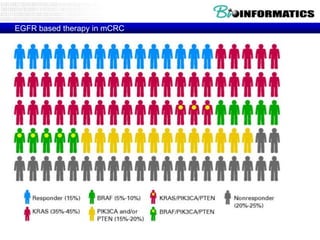
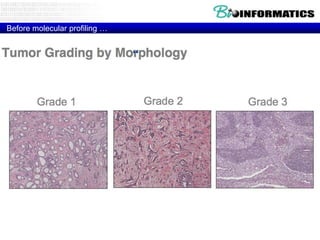
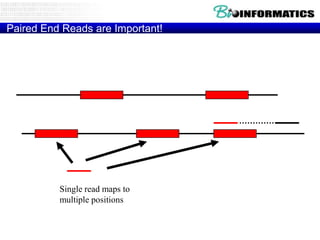
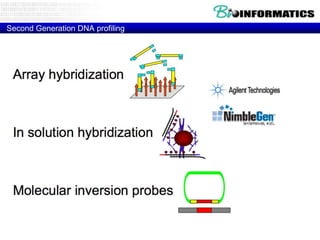
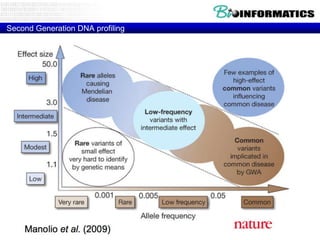
The document provides an overview of genomics and molecular profiling techniques. It discusses:
- The lab for bioinformatics and computational genomics which has 10 "genome hackers" and 42 scientists.
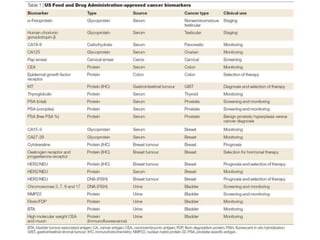
- An introduction to personalized medicine and biomarkers.
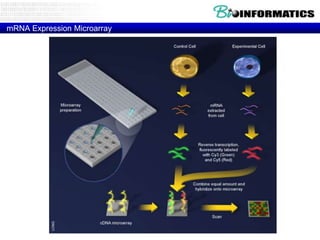
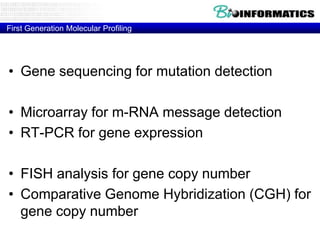
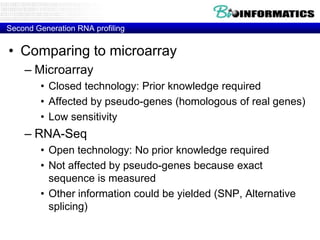
- First generation molecular profiling techniques like gene sequencing, microarrays, PCR.
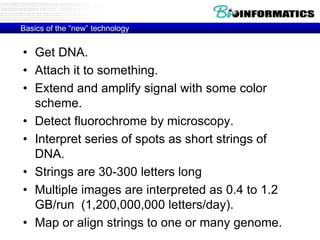
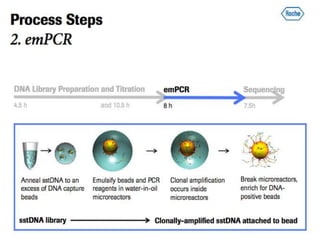
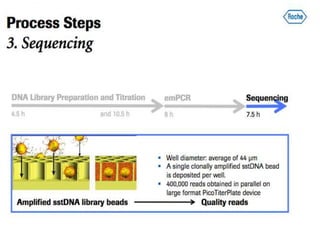
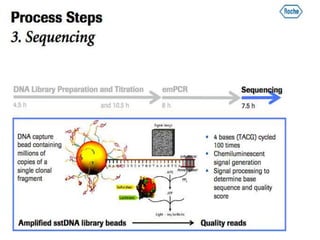
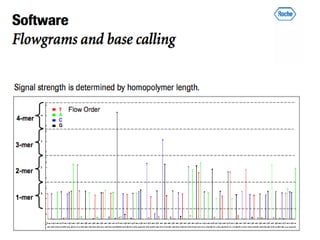
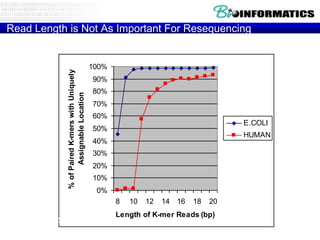
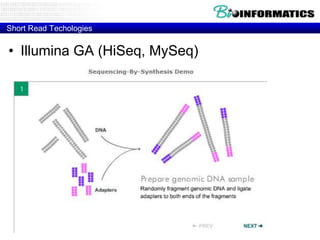
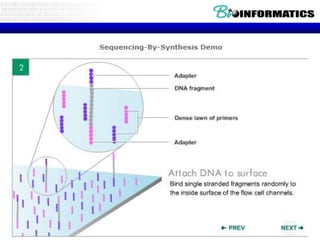
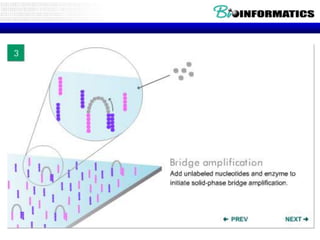
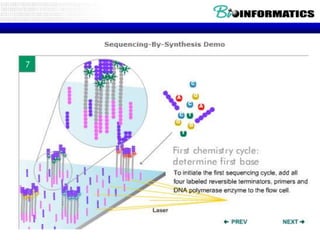
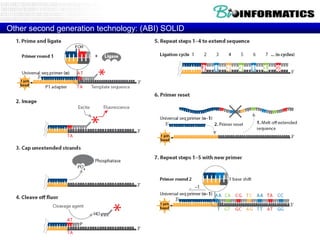
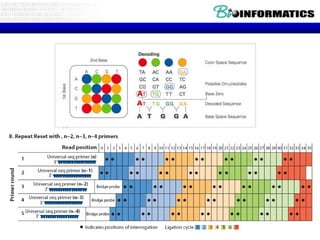
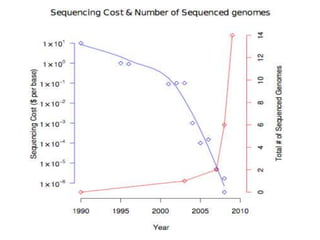
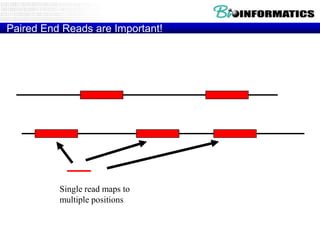
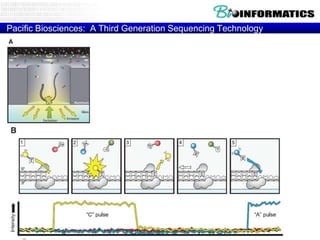
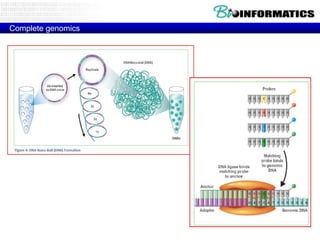
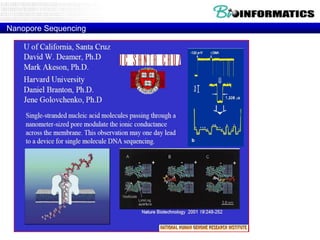
- Next generation sequencing techniques like Roche 454, Illumina, SOLID which allow high throughput sequencing.
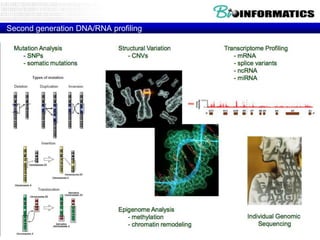
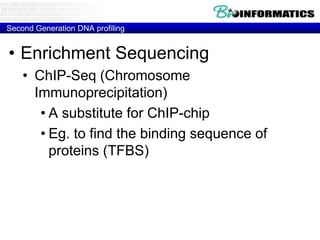
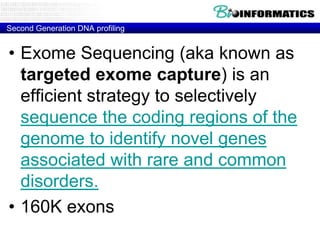
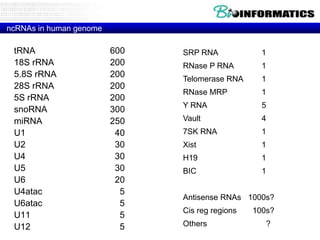
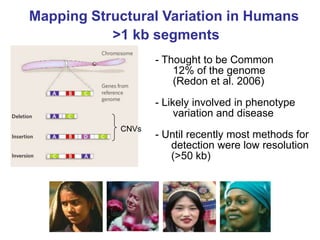
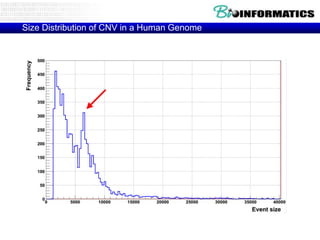
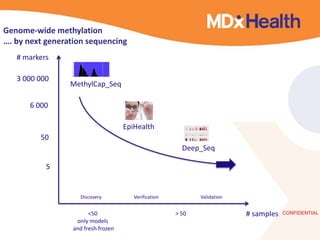
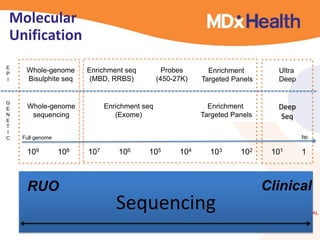
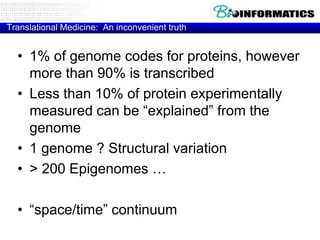
- Next generation applications like RNA sequencing, exome sequencing, epigenetic profiling.
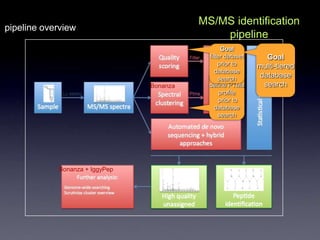
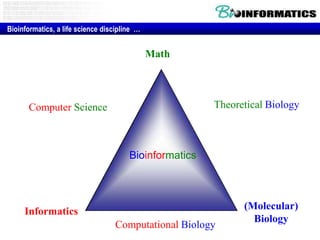
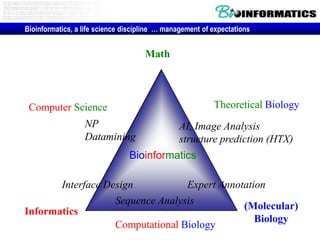
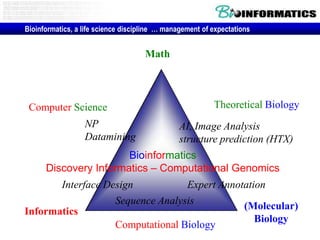
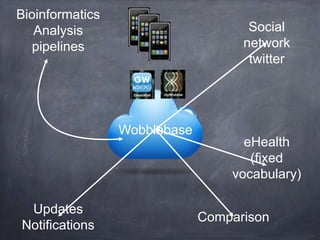
- The role of bioinformatics in analyzing large genomic and molecular profiling data.