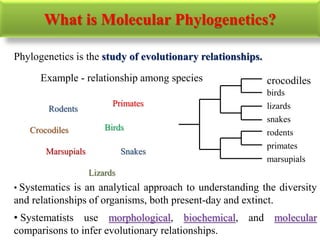
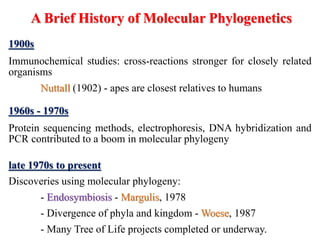
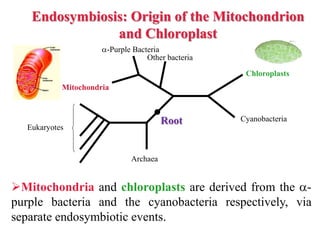
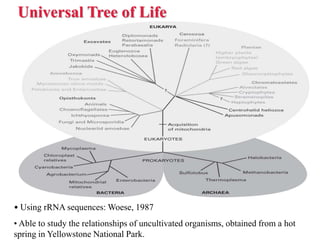
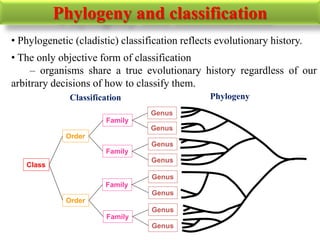
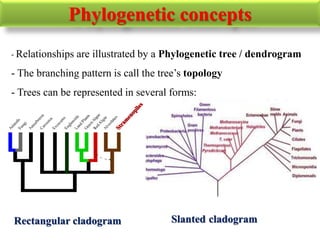
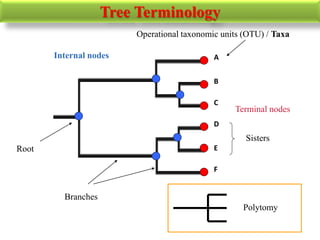
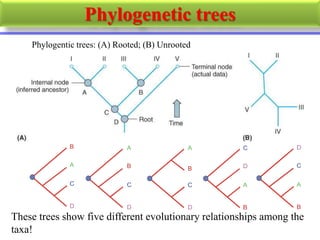
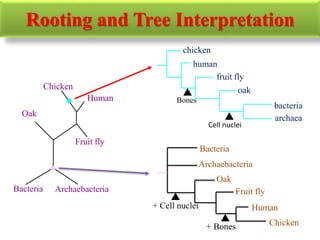
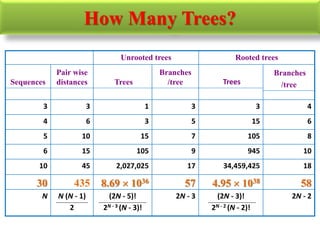
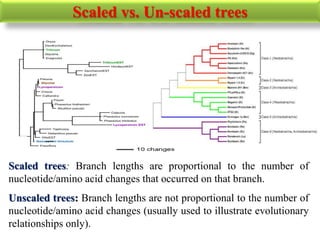
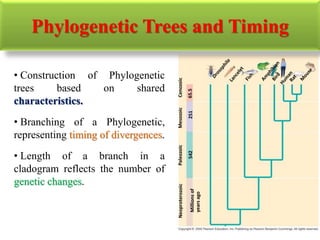
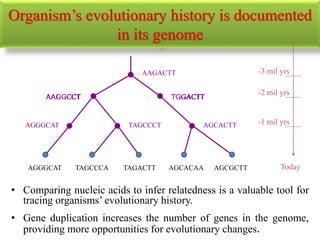
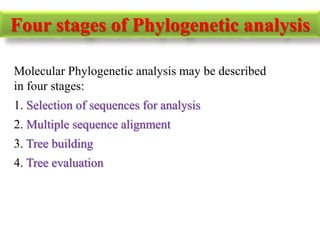
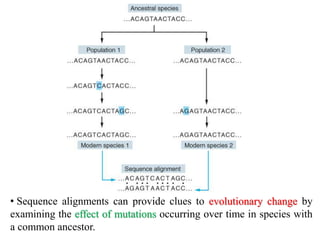
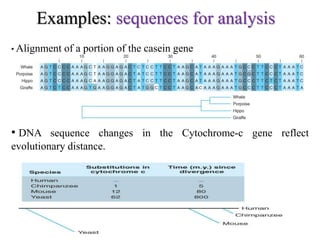
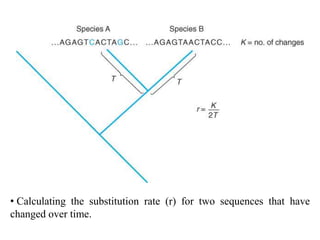
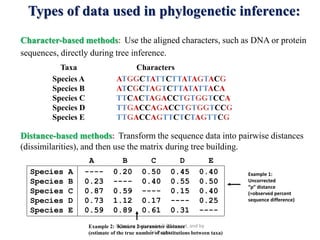
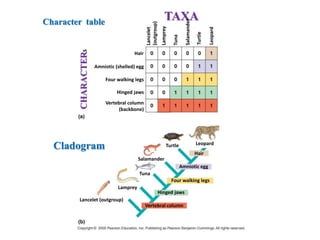
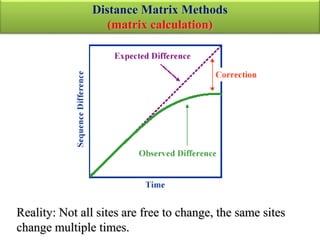
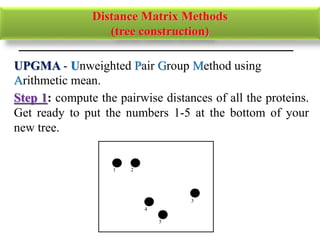
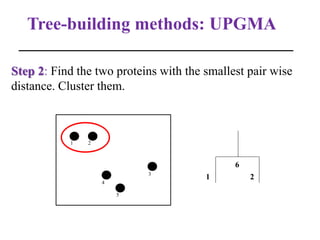
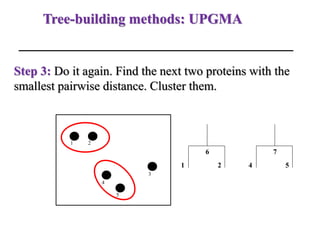
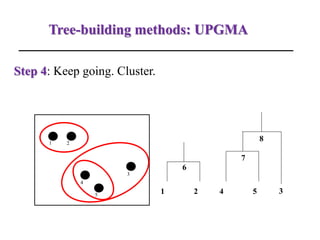
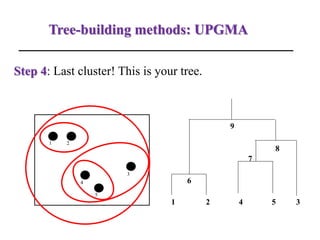
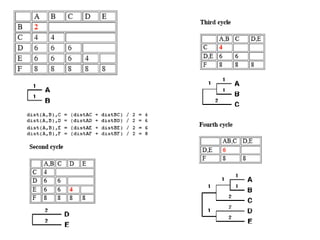
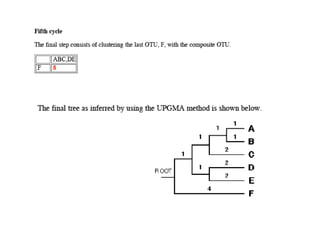
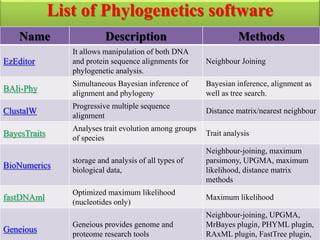
The document presents an overview of molecular phylogenetics, which is the study of evolutionary relationships among organisms using molecular data. It discusses the history, methods, and applications of phylogenetic analysis, including tree construction and data types used for inferring evolutionary relationships. Additionally, it highlights various fields that benefit from phylogenetic insights, such as conservation, medicine, and forensic investigations.