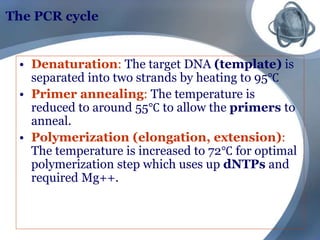
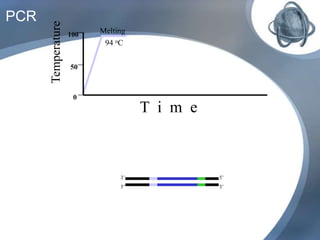
The document outlines the principles and processes of three genetic techniques: Polymerase Chain Reaction (PCR), Random Amplified Polymorphic DNA (RAPD), and Restriction Fragment Length Polymorphism (RFLP). It provides detailed information on PCR methodology, including temperature cycles and primer design, as well as how RAPD uses short primers to generate genetic markers. Additionally, it discusses RFLP's reliance on restriction enzymes for detecting genetic variations through electrophoresis and hybridization techniques.