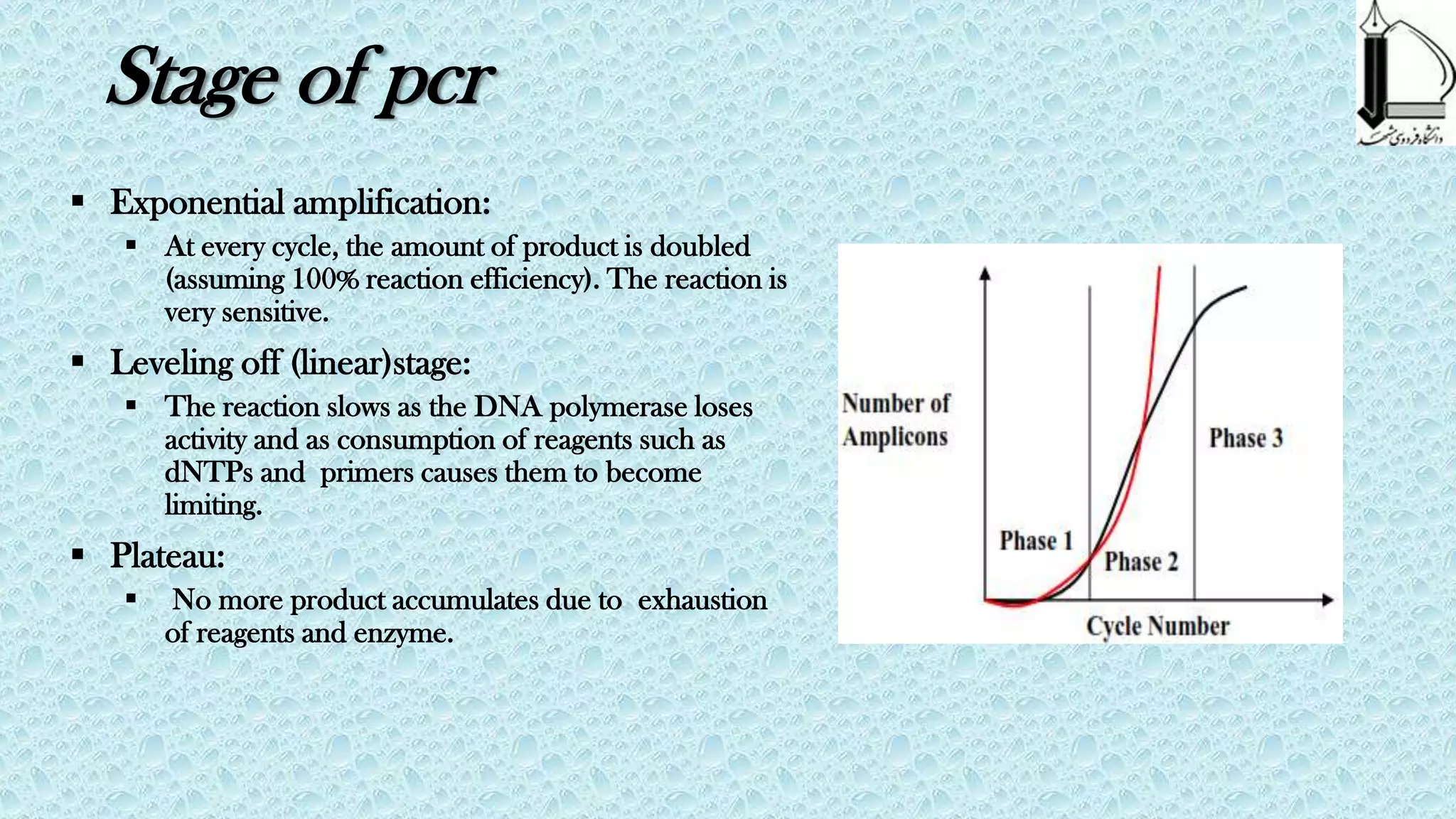
PCR was invented in 1983 by Kary Mullis and revolutionized molecular biology. It allows for the exponential amplification of specific DNA regions using thermal cycling and two primers. Key developments included the use of Taq polymerase, automation using thermal cyclers, and optimization of components and cycling conditions. Variations like nested PCR, hot start PCR, and RT-PCR expanded PCR applications. PCR provides a powerful and sensitive technique for detecting and analyzing DNA and RNA.