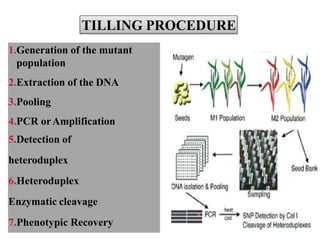
The document discusses the technique of TILLING (Targeting Induced Local Lesions in Genomes), a reverse genetics approach that combines chemical mutagenesis with PCR screening to identify point mutations in specific genes. It outlines the TILLING process, its applications in functional genomics and genetic engineering, as well as the advantages and limitations of the method. Additionally, it introduces eco-tilling, a variation that focuses on identifying natural genetic variation, highlighting its potential in crop improvement.