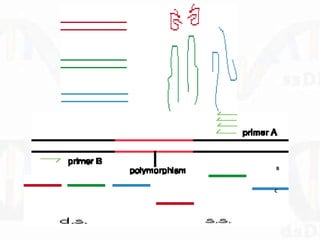
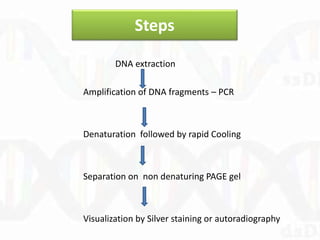
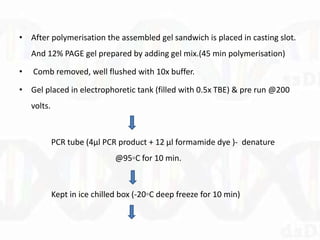
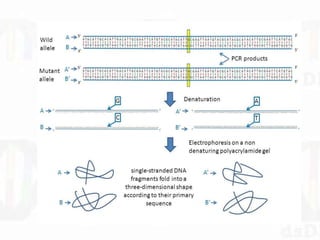
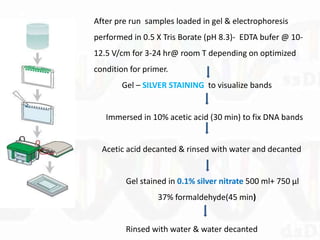
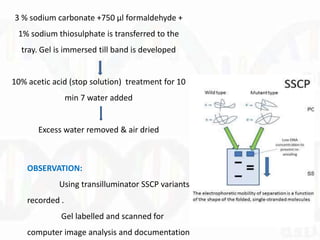
Single-strand conformation polymorphism (SSCP) is a technique that detects variations in single-stranded DNA sequences. It involves PCR amplification of a target region, denaturing the PCR products to generate single strands, and separating the single strands on a non-denaturing gel based on differences in electrophoretic mobility caused by variations in nucleotide sequence. This allows sequences to be distinguished and variants detected without sequencing. SSCP is useful for discovering new polymorphisms and detecting mutations for diagnostic applications.