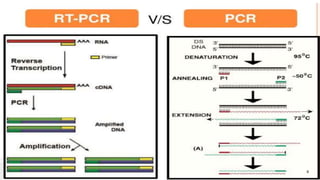
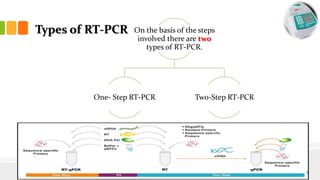
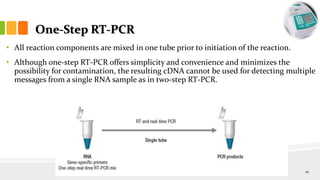
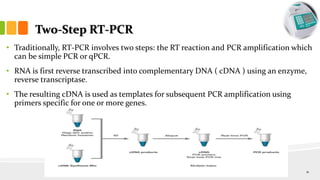
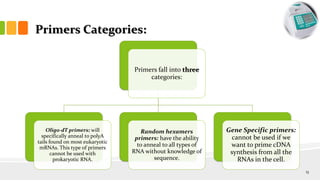
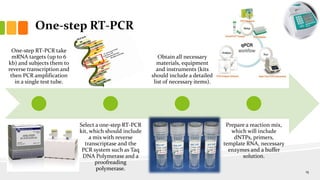
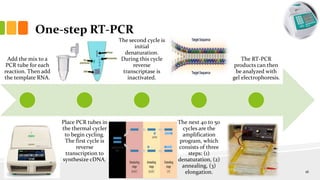
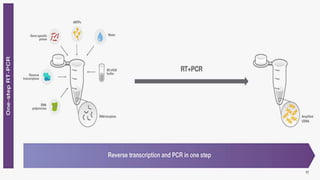
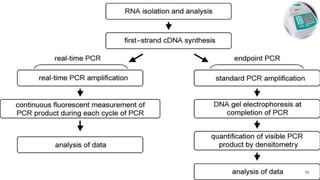
The document discusses reverse transcription polymerase chain reaction (RT-PCR) techniques, outlining the differences between conventional PCR and RT-PCR, particularly focusing on their application for detecting RNA expression and gene quantification. It explains one-step and two-step RT-PCR methods, their protocols, and details on primer types, emphasizing the importance of high-quality RNA and the need for controls to avoid DNA contamination. Additionally, it covers the diverse applications of RT-PCR in research, genetic disease diagnosis, cancer detection, and gene expression analysis.