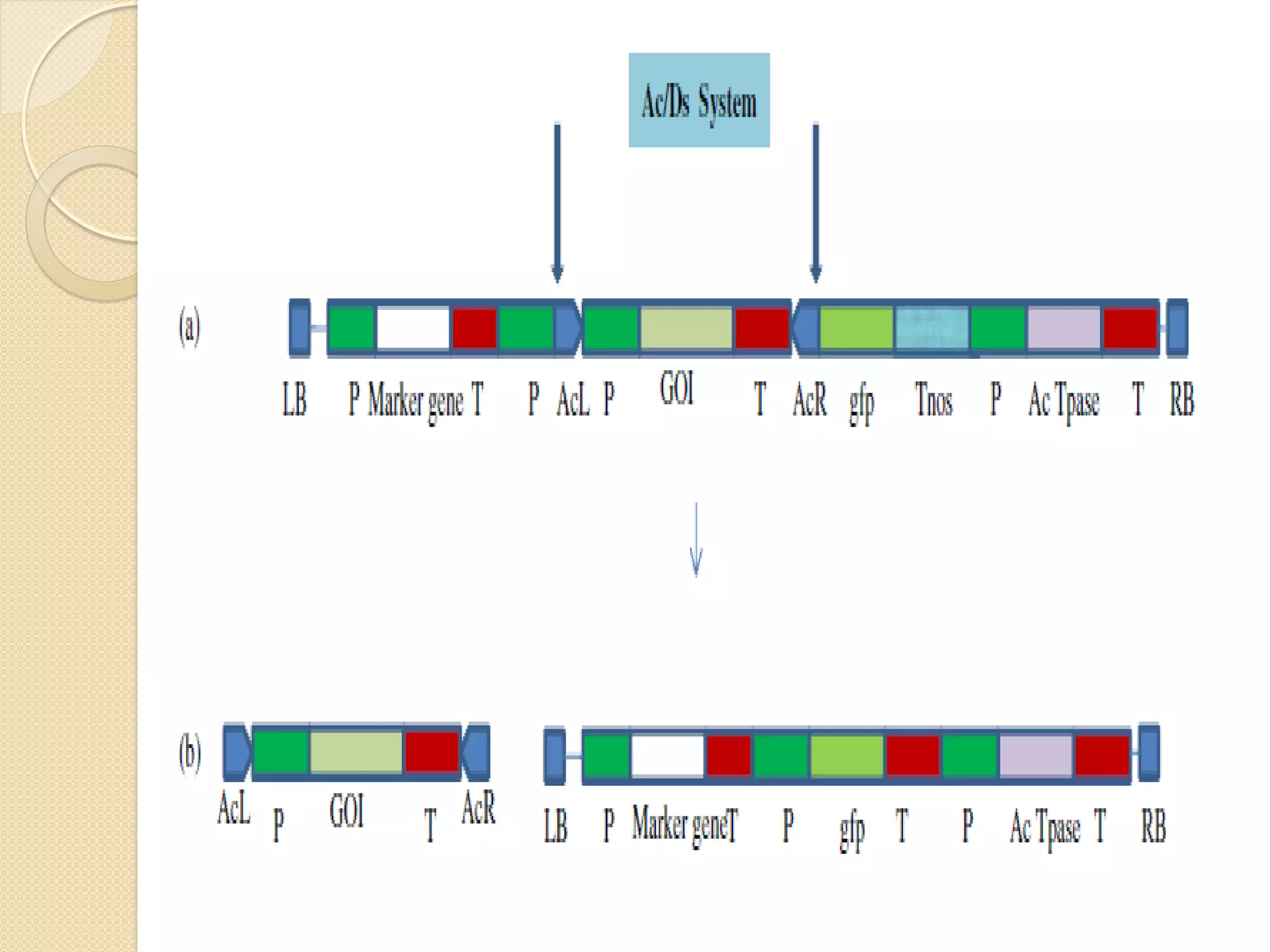
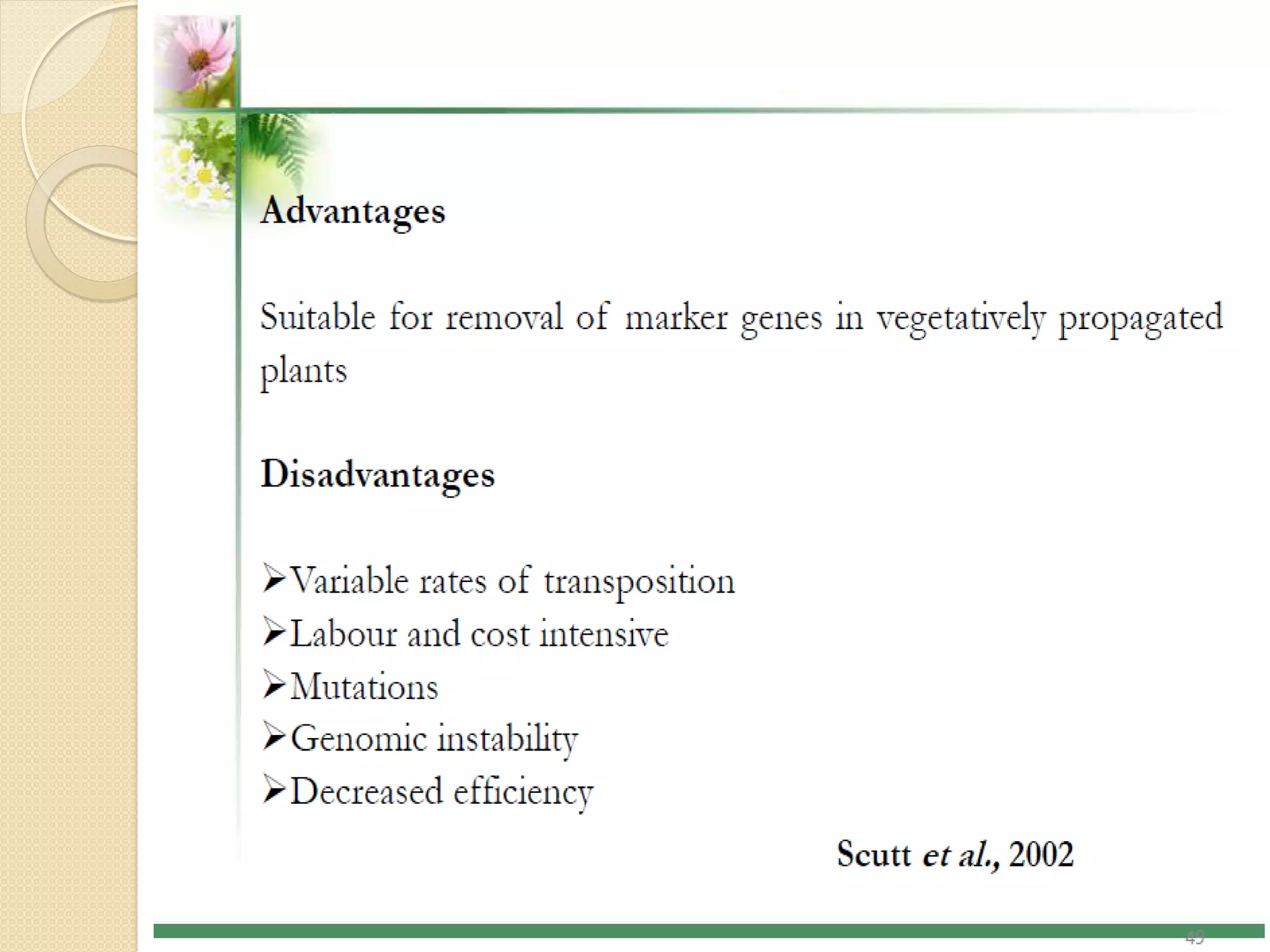
This presentation discusses strategies for developing transgenic plants without selectable marker genes. Marker genes are commonly used to identify transformed cells but can be problematic for public acceptance and future transformations. Methods described for producing marker-free transgenics include the MAT system which uses oncogenes for selection instead of antibiotics, site-specific recombination systems which flank the marker gene for later excision, and transposon-based systems which separate the gene of interest from the marker gene. While several viable methods exist, more work is still needed before marker-free crops can be commercialized. Removing marker genes supports multiple-gene stacking and improves public acceptance of transgenic technologies.












![MAT SYSTEM
A positive selection system
Unique as it uses morphological changes caused by
oncogene [ipt gene] or rhizogene (the rol gene) of A.
Tumefaciens which control the endogenous levels of plant
hormones and the cell responses to PGR as the selection
marker.
A chosen GOI is placed adjacent to a multigenic element
flanked by RS recombination sites. A copy of the selectable
ipt gene from A.tumefaciens is inserted between these sites.](https://image.slidesharecdn.com/markerfreetransgenicstrategy-190519082754/75/Marker-free-transgenic-strategy-17-2048.jpg)