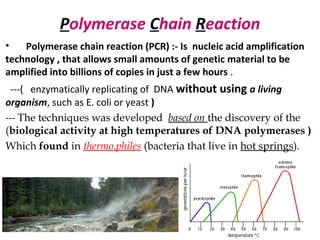
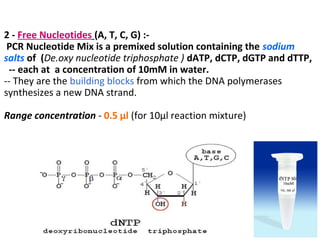
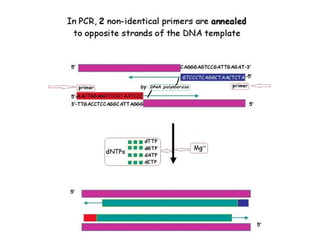
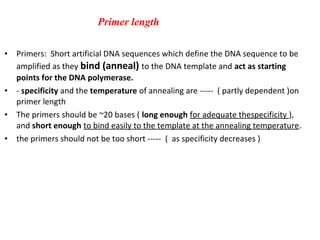
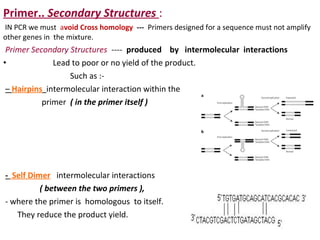
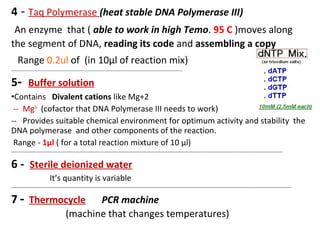
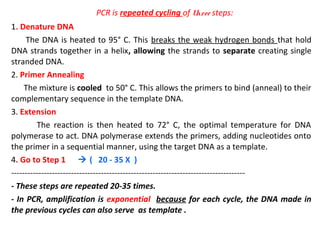
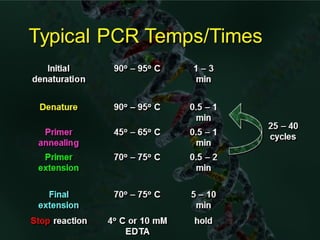
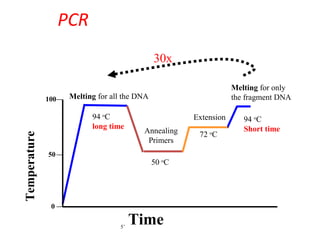
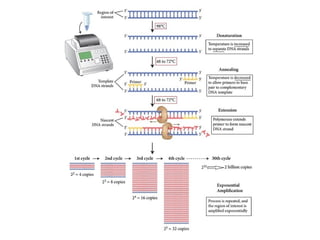
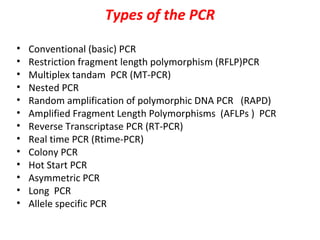
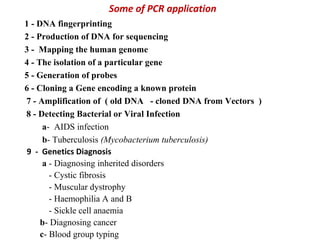
Polymerase chain reaction (PCR) is a technique used to amplify small segments of DNA across several orders of magnitude. PCR uses DNA polymerase to replicate a target DNA segment using primers that flank the region of interest. The reaction involves cycling between high and low temperatures to denature the DNA, anneal primers, and extend new strands. Kary Mullis developed PCR in 1983, allowing for rapid amplification of DNA without using living cells. PCR has many applications, including DNA fingerprinting, sequencing, genetic testing, and detecting bacterial/viral infections.