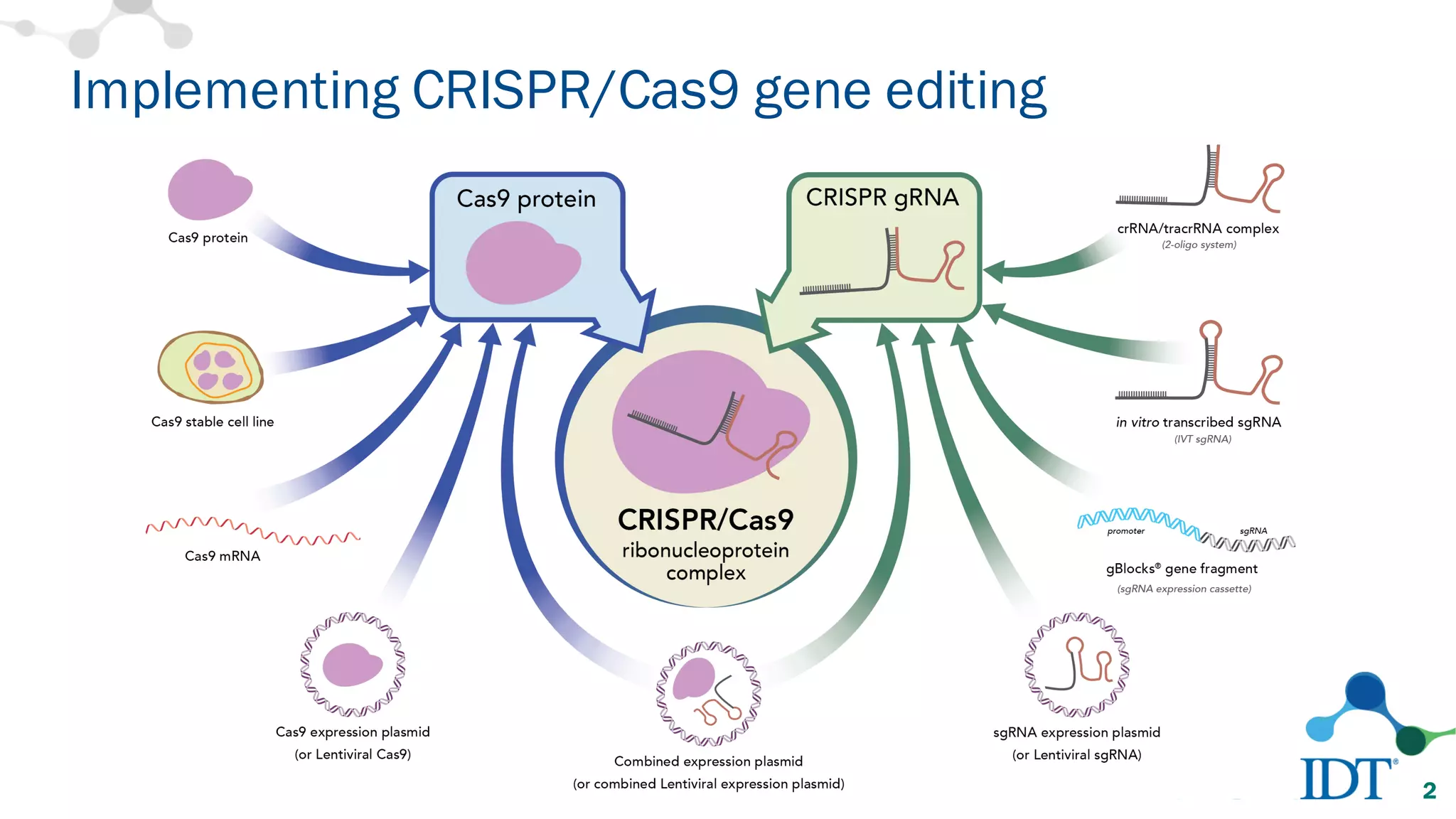
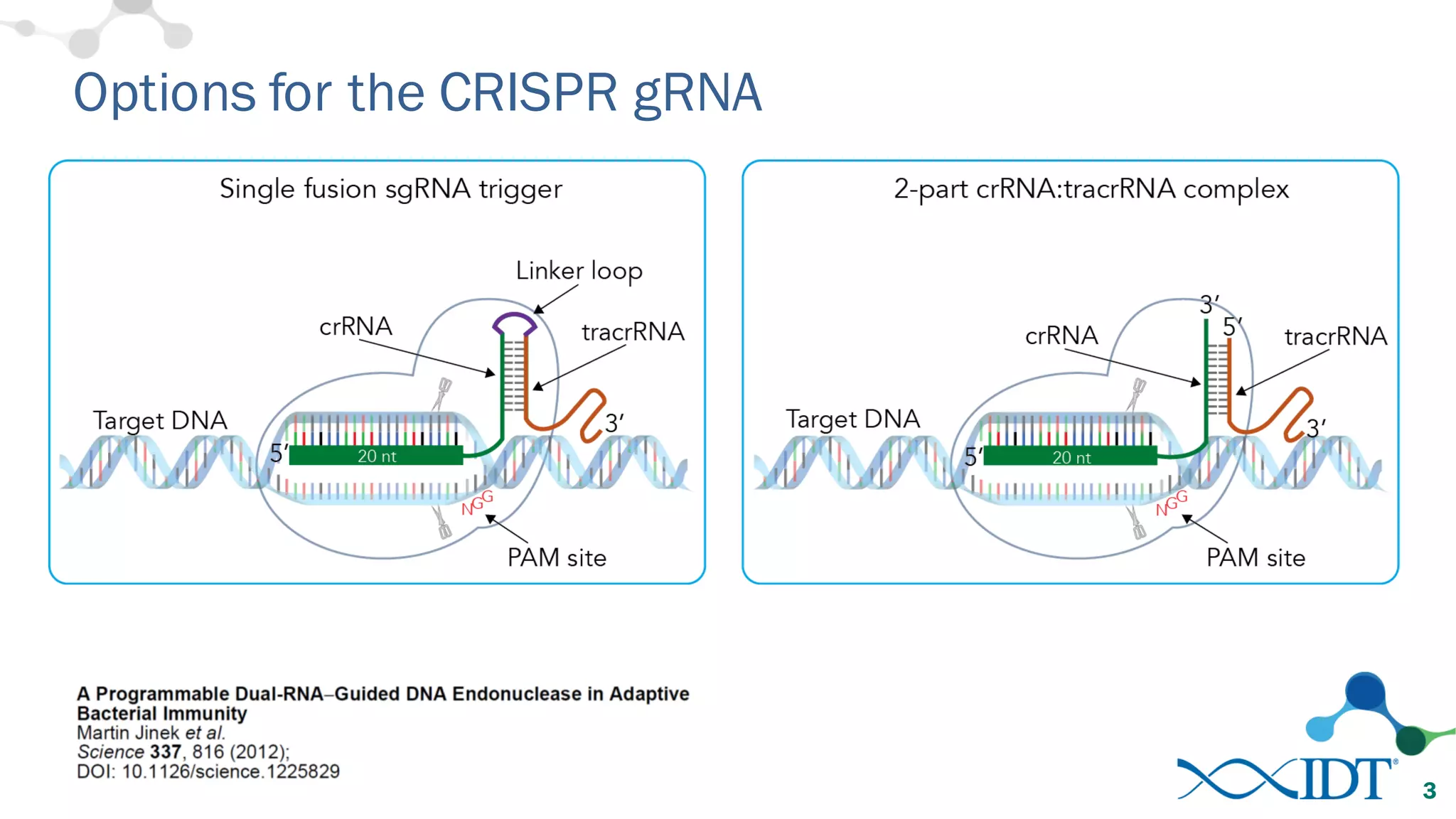
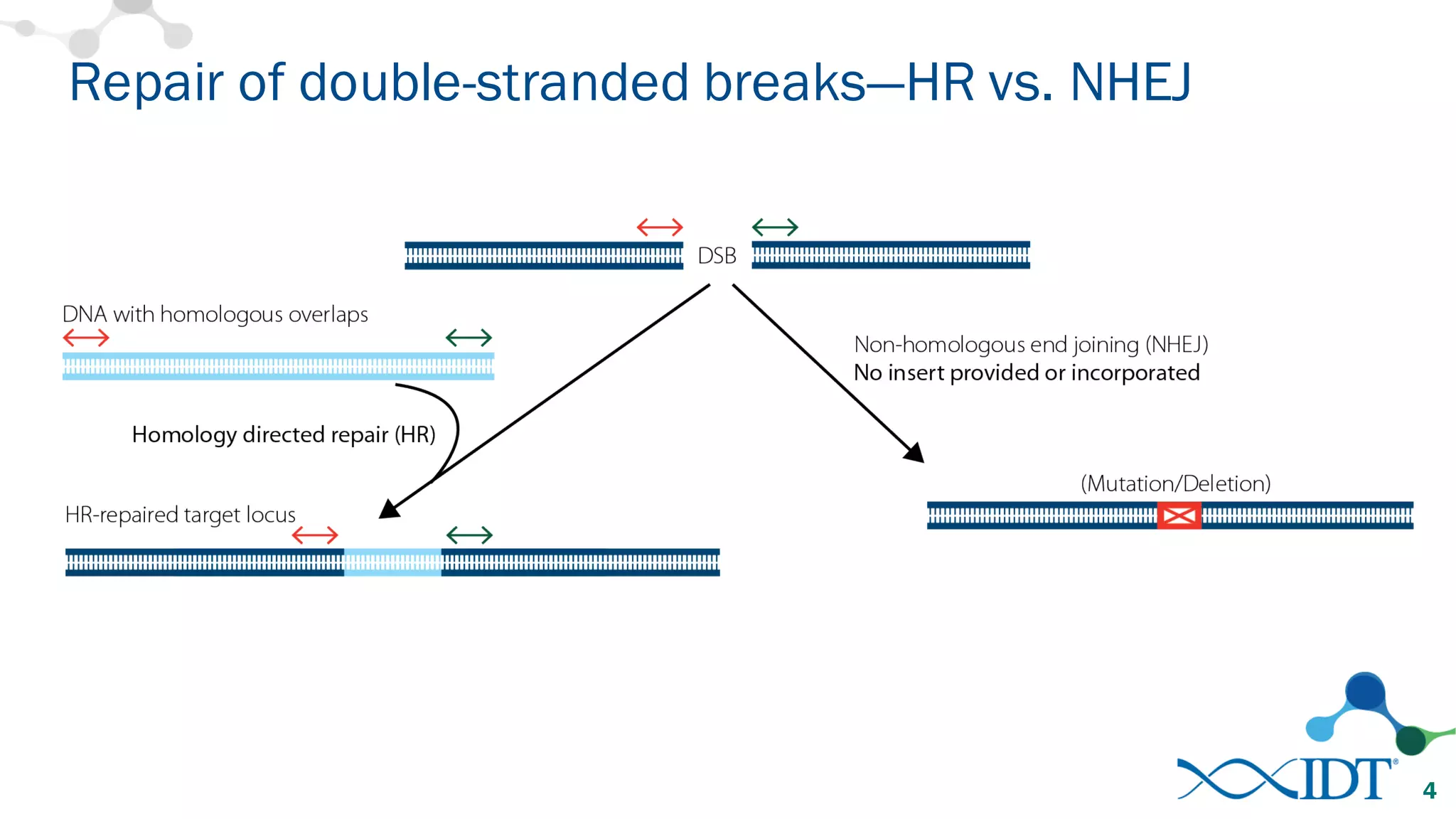
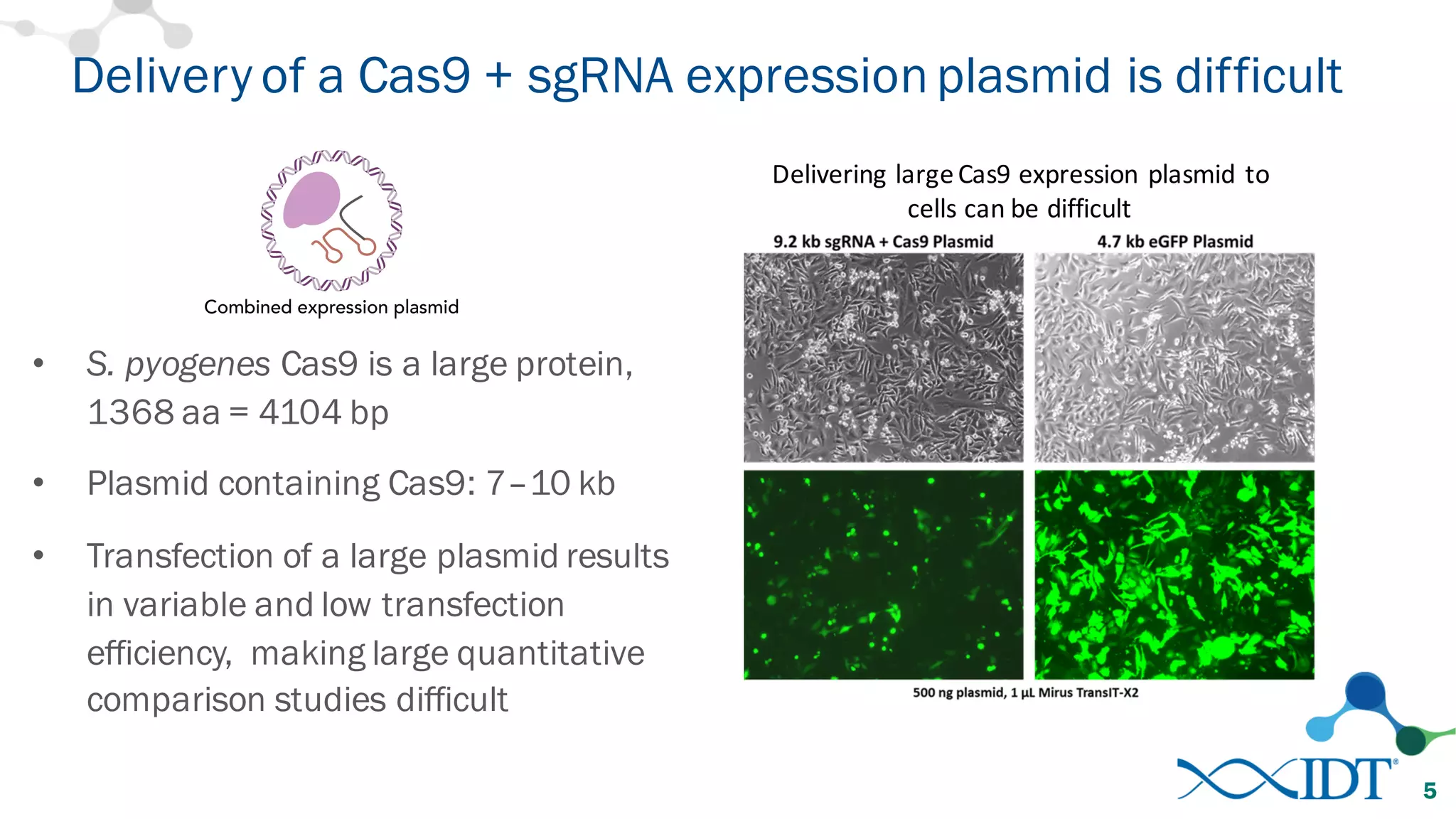
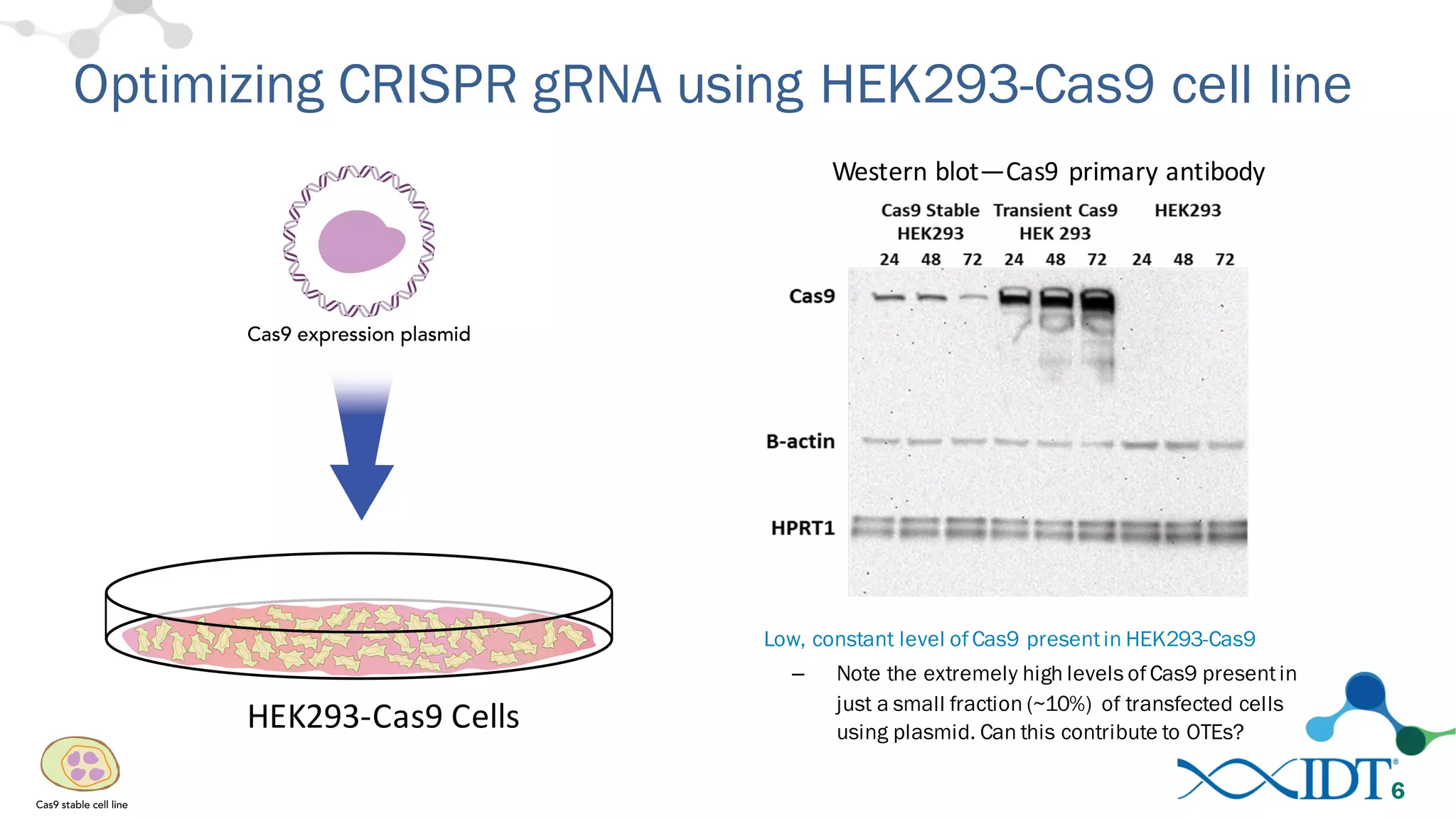
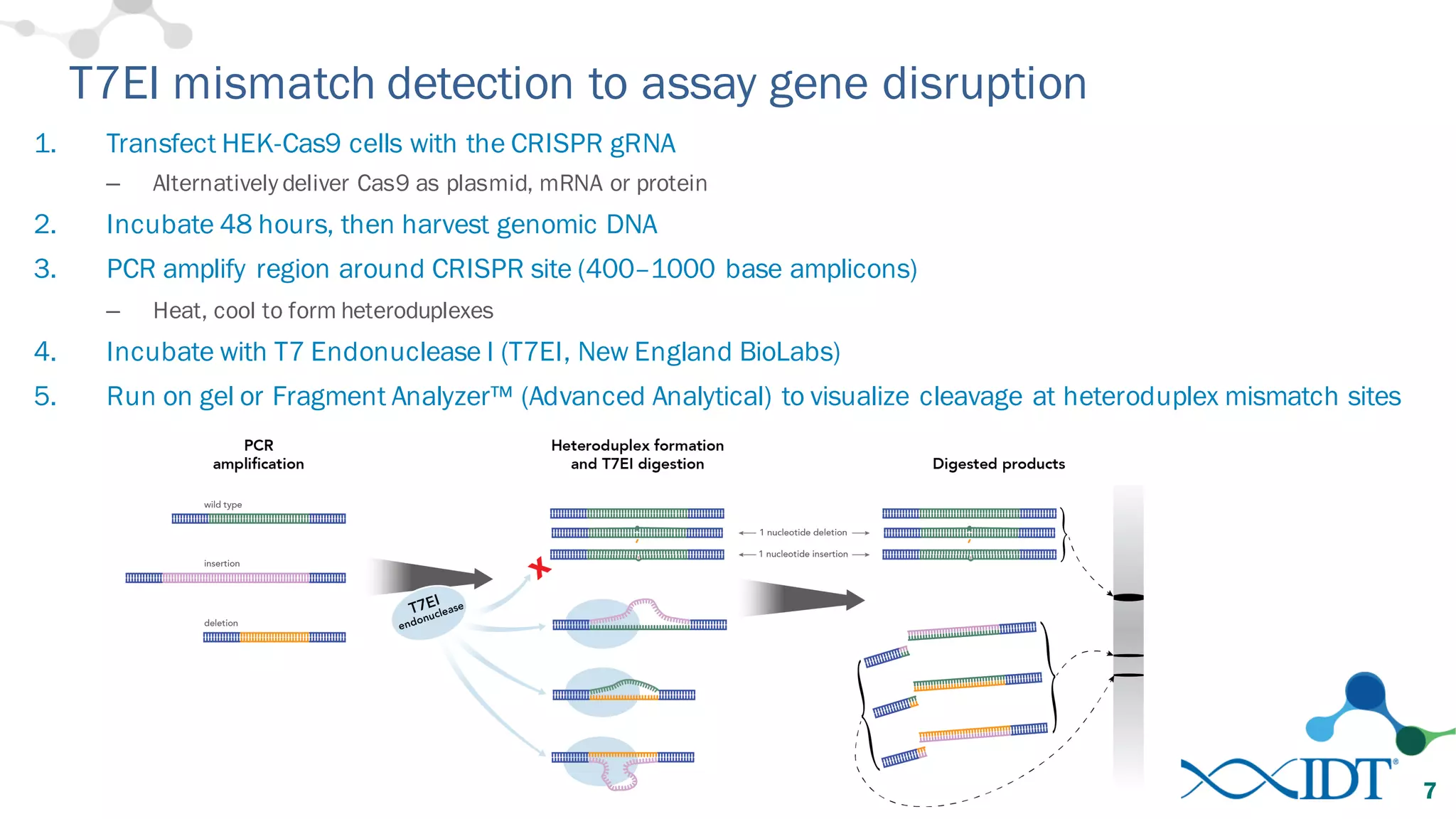
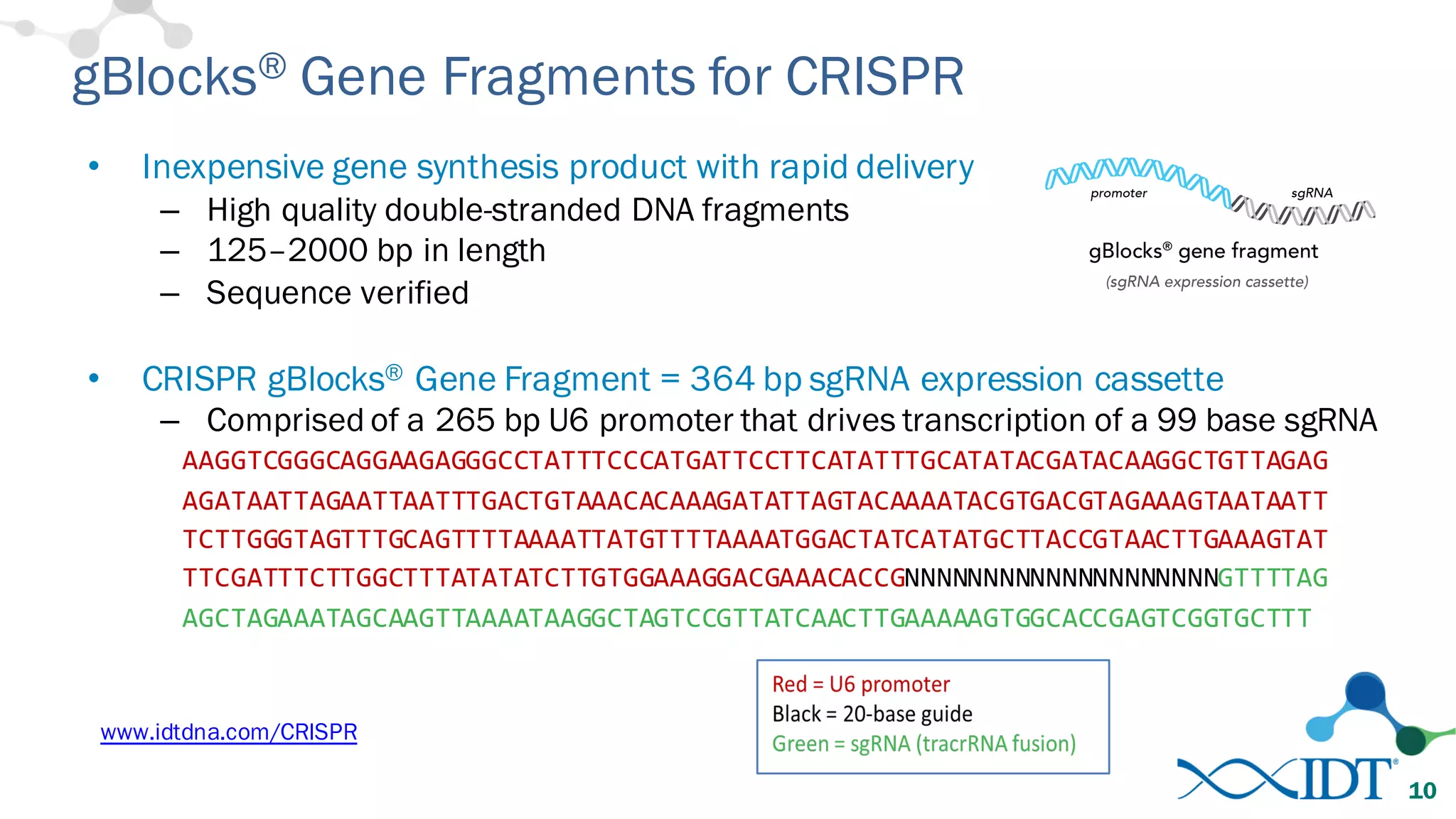
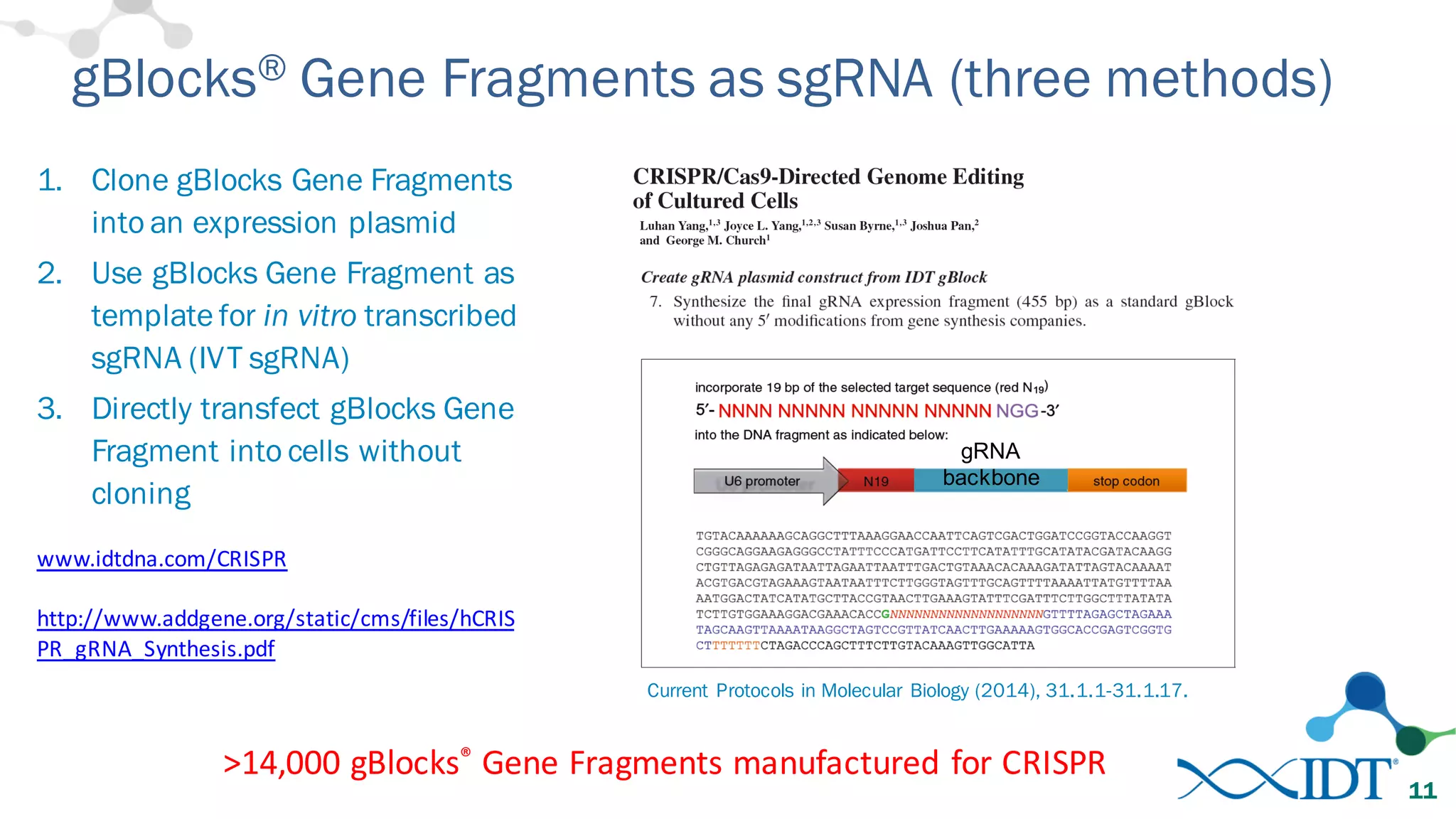
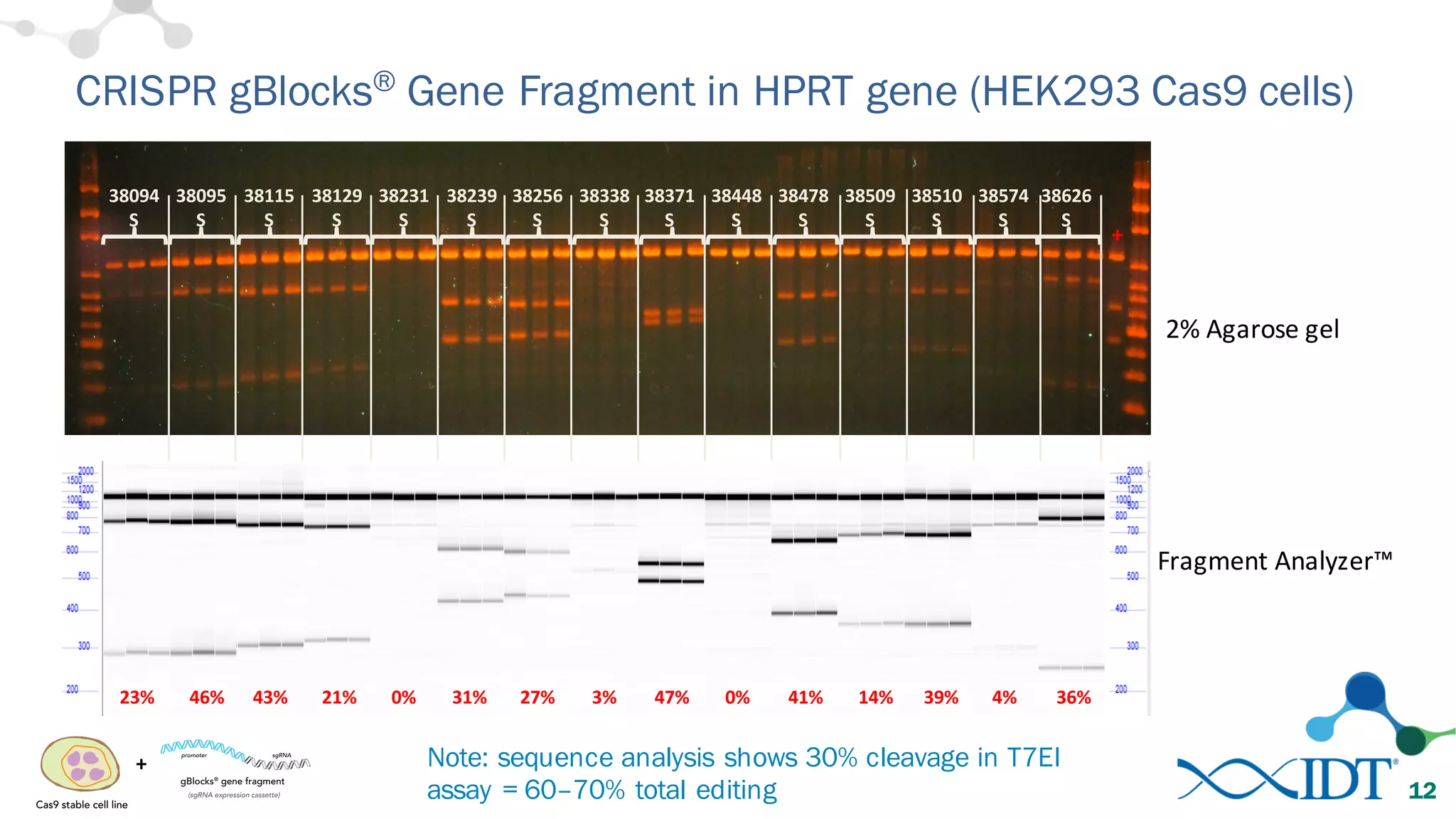
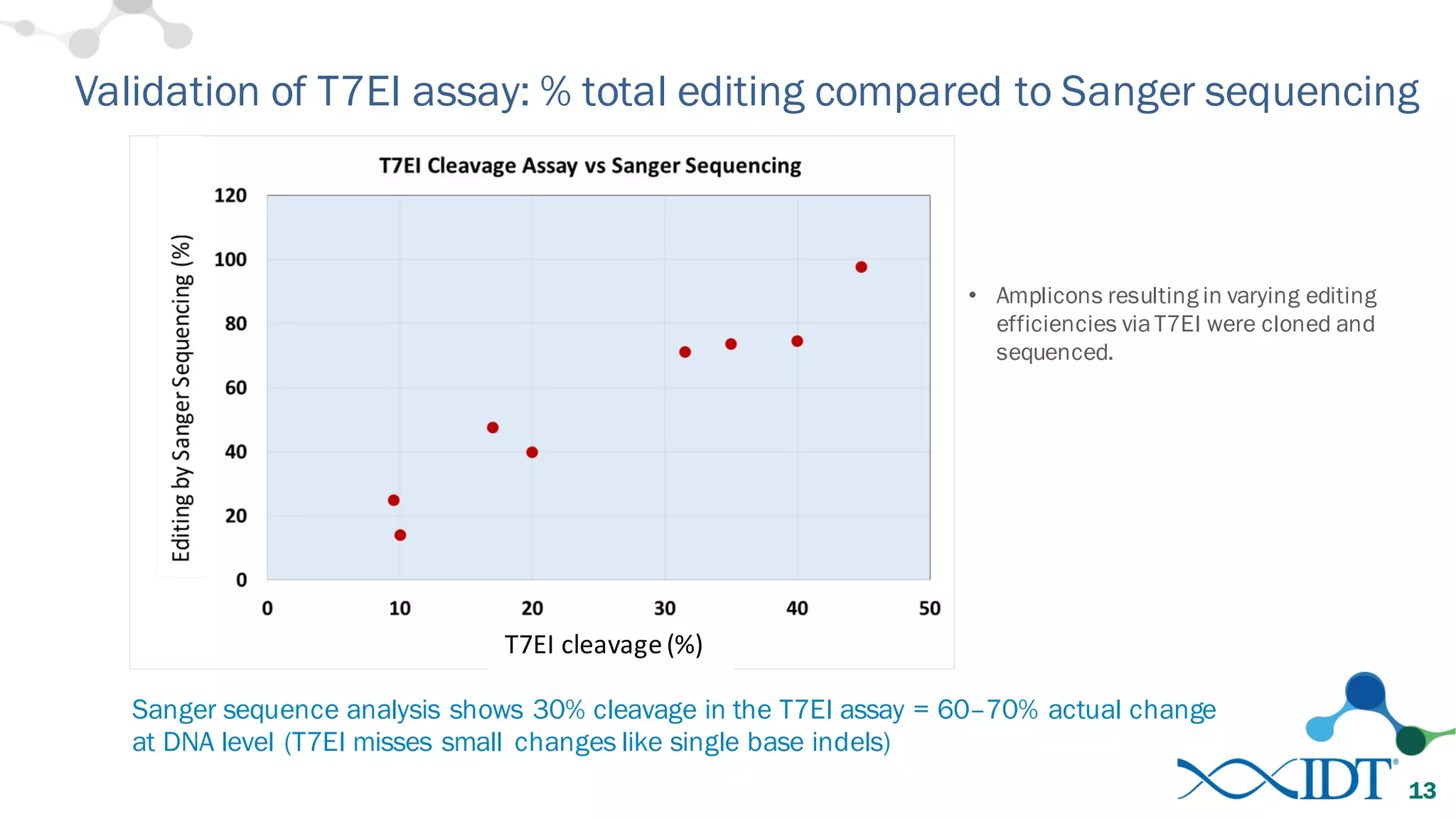
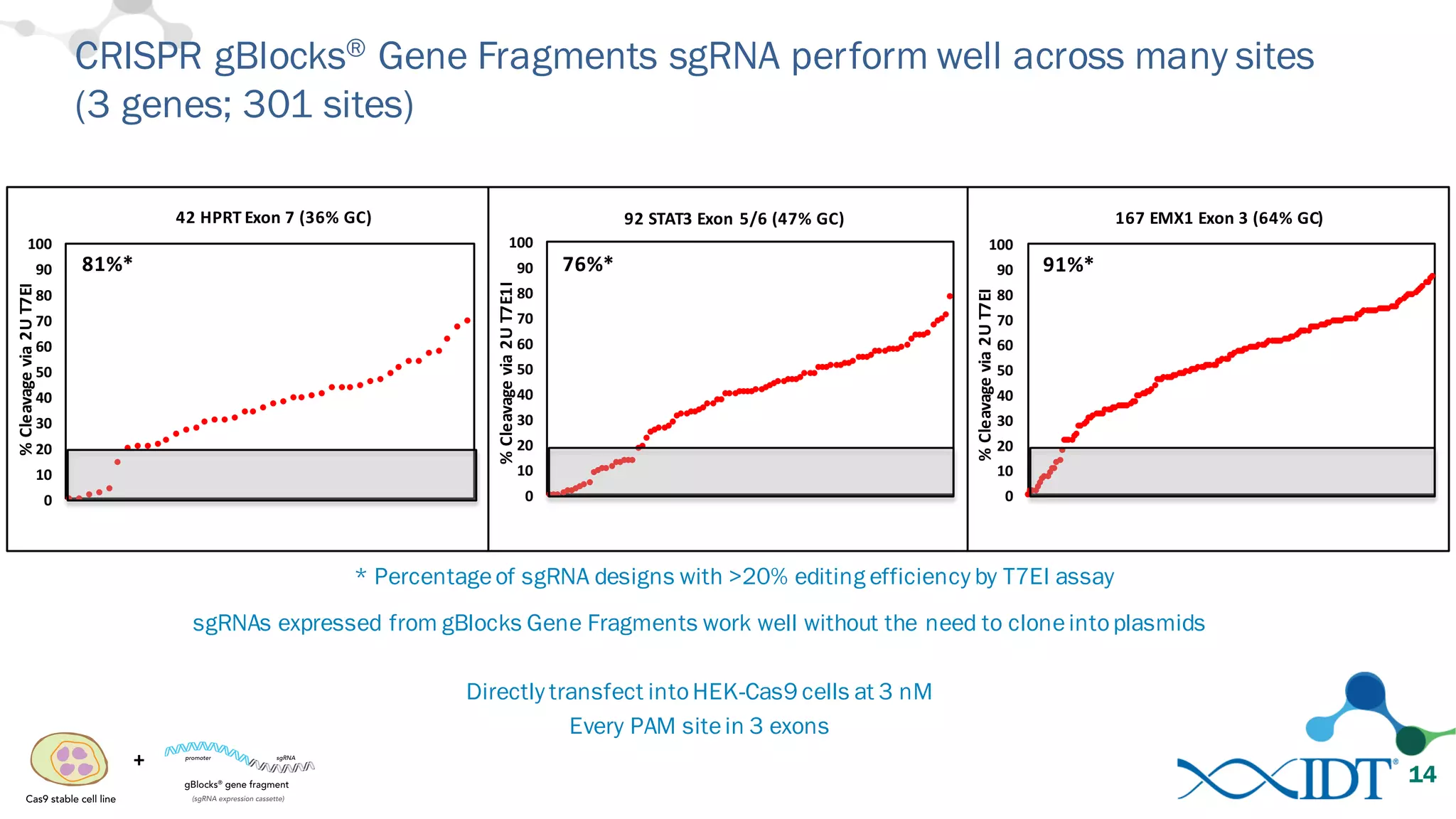
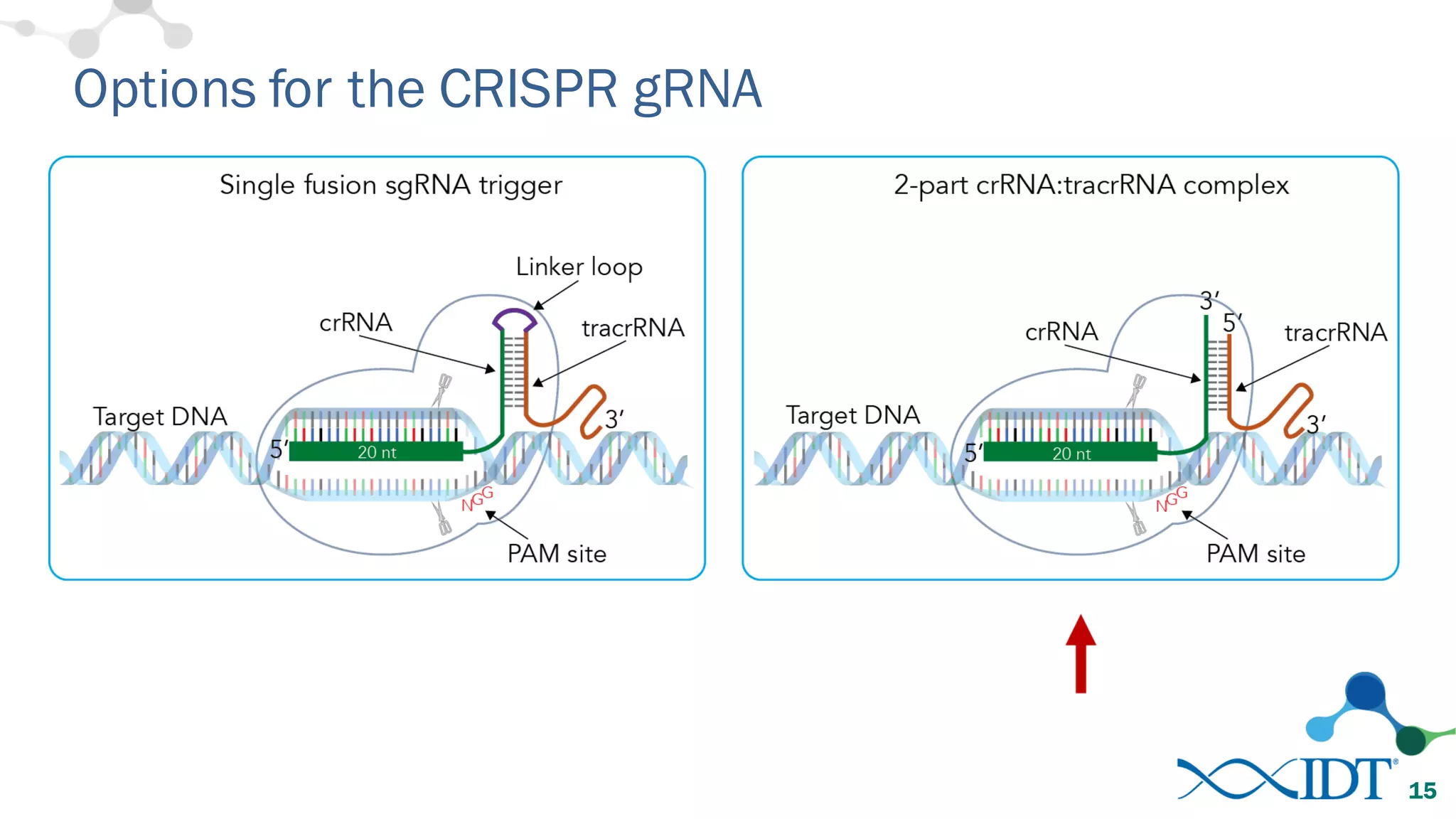
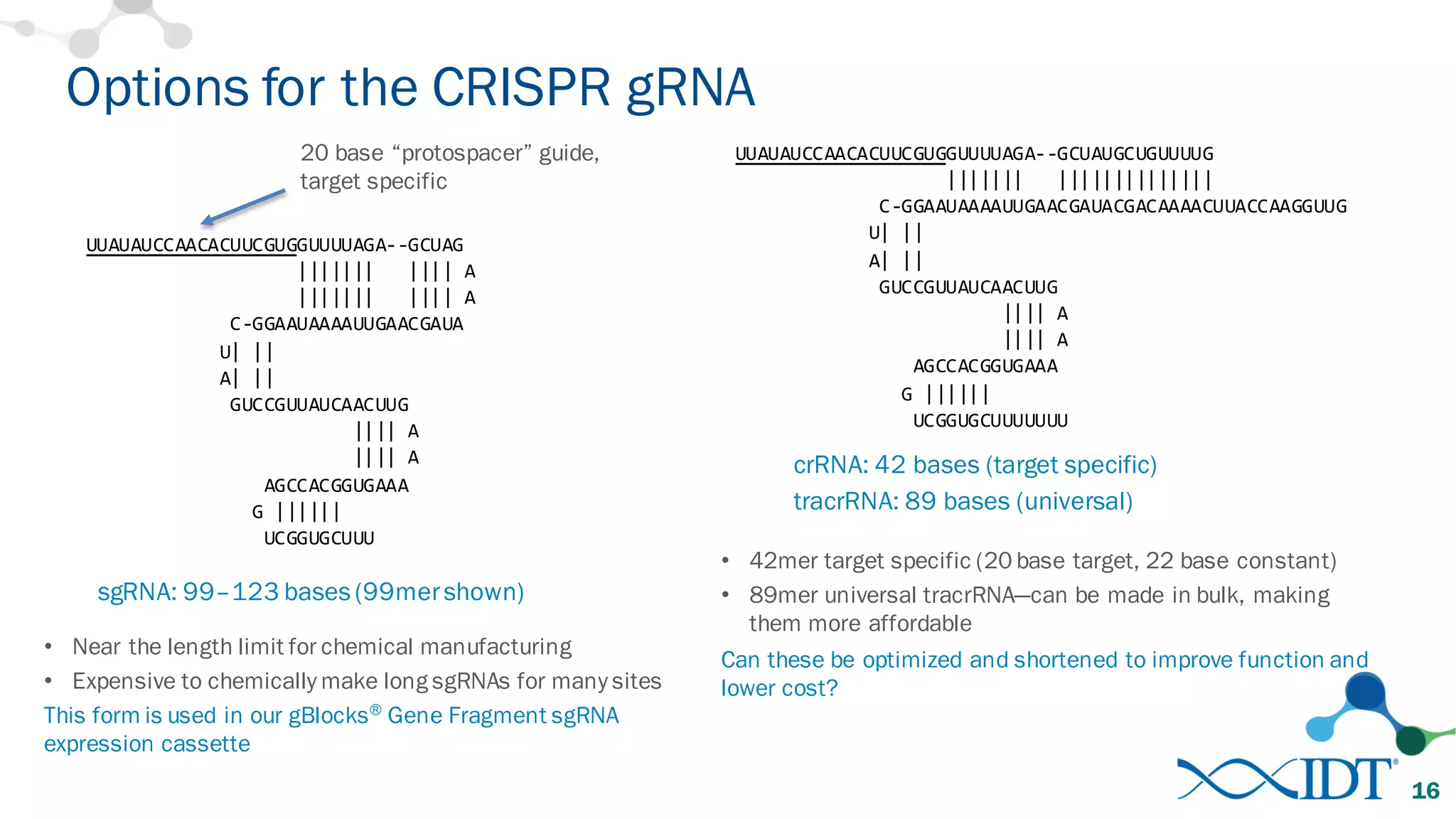
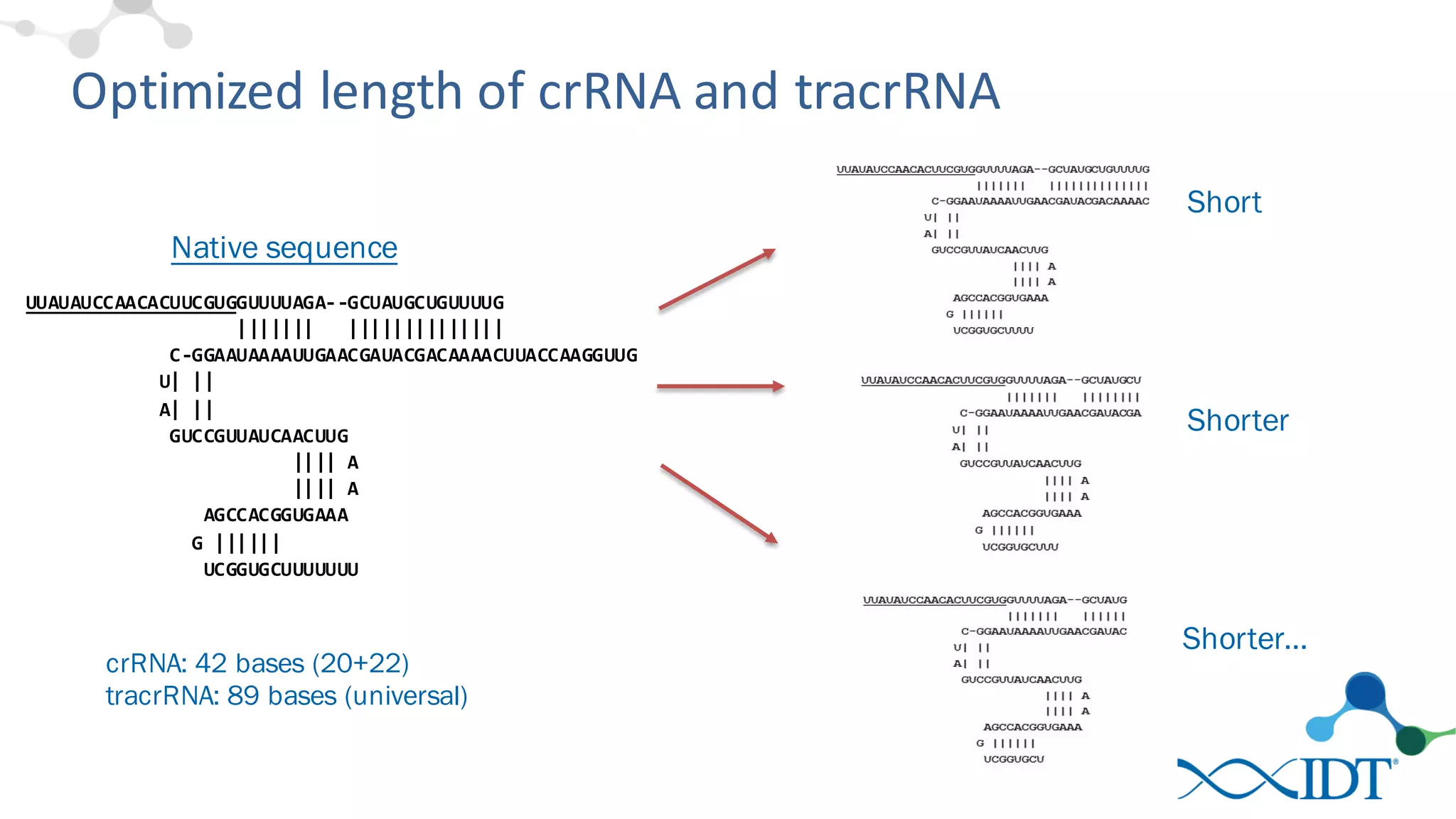
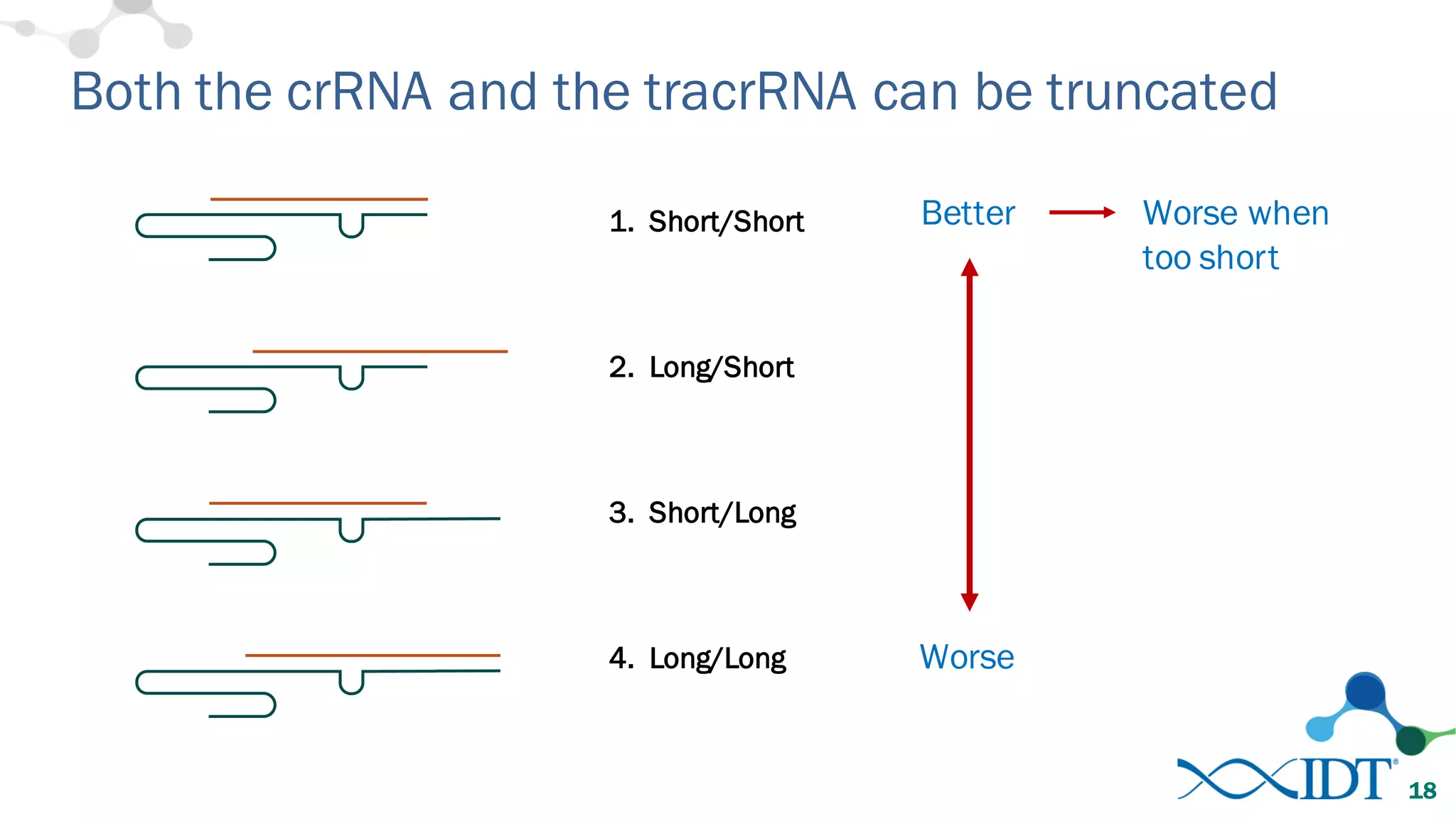
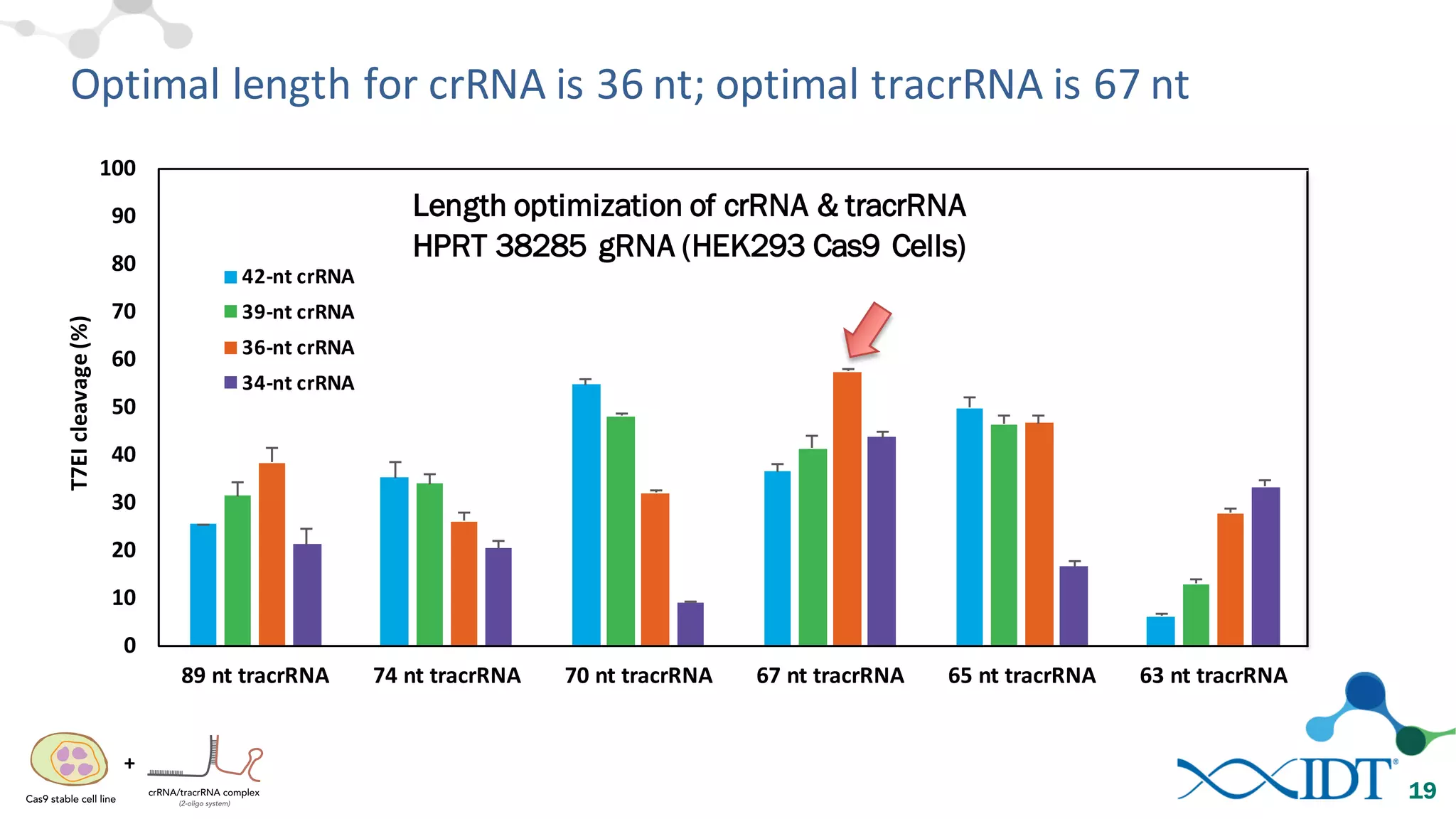
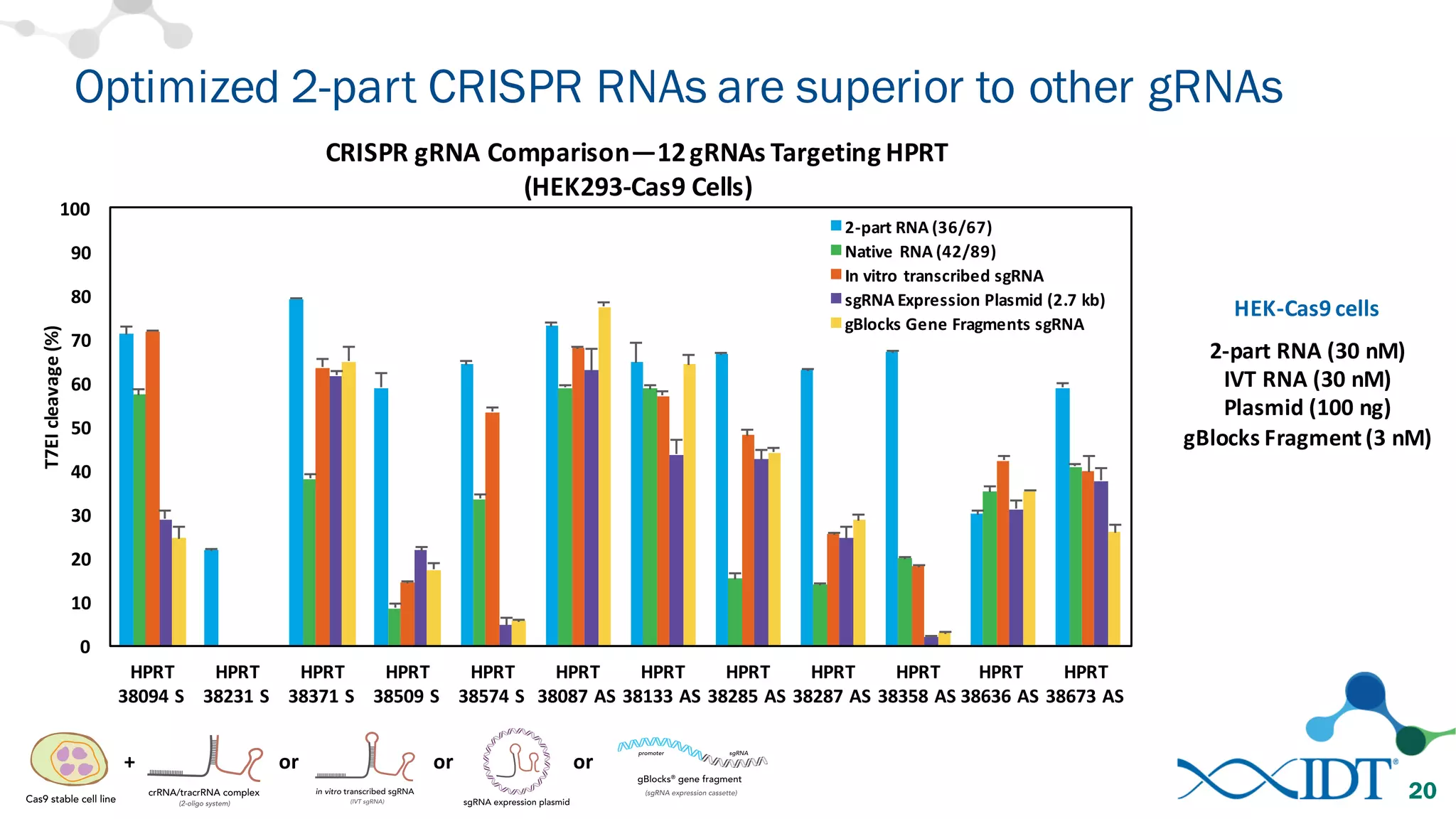
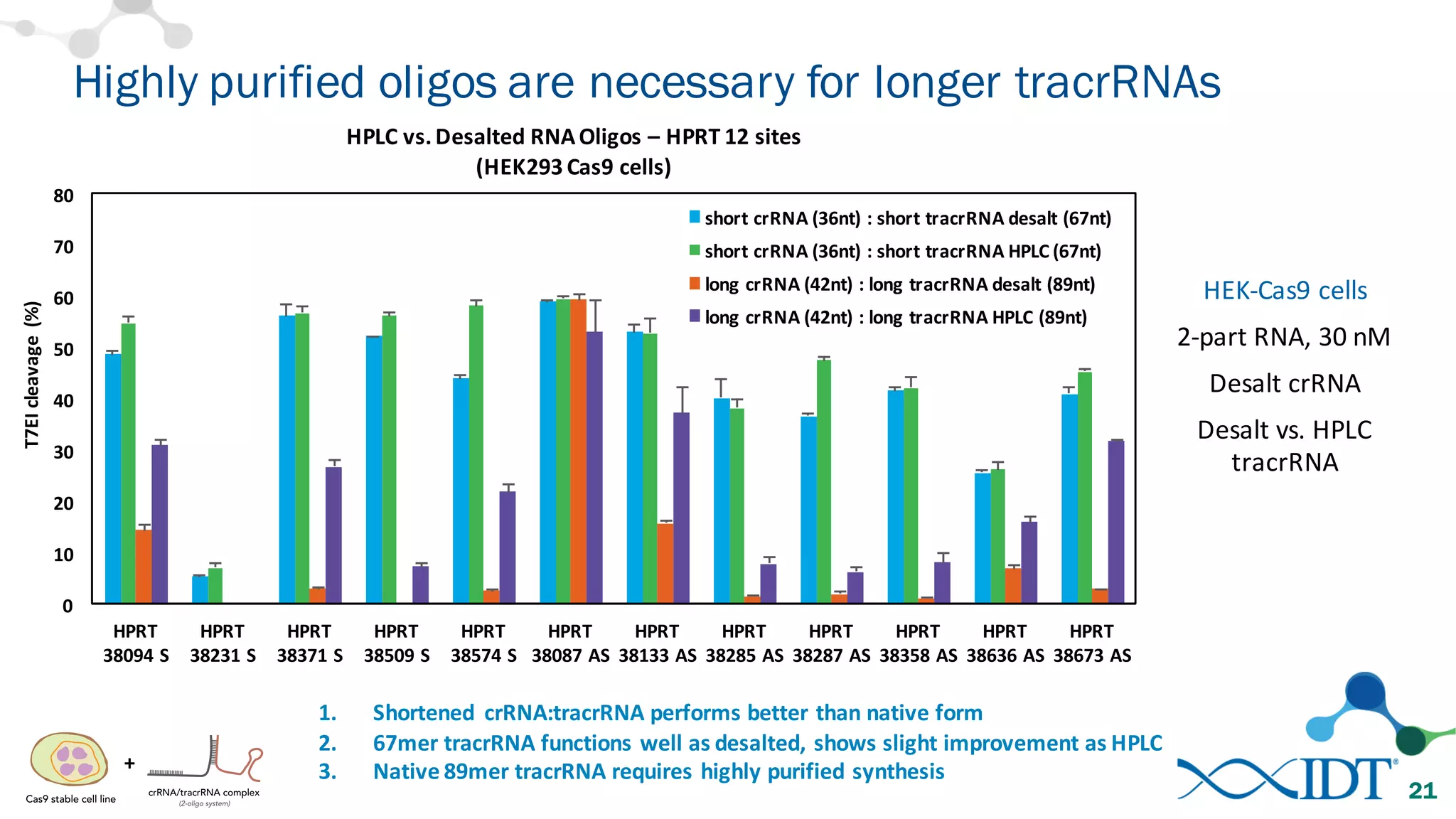
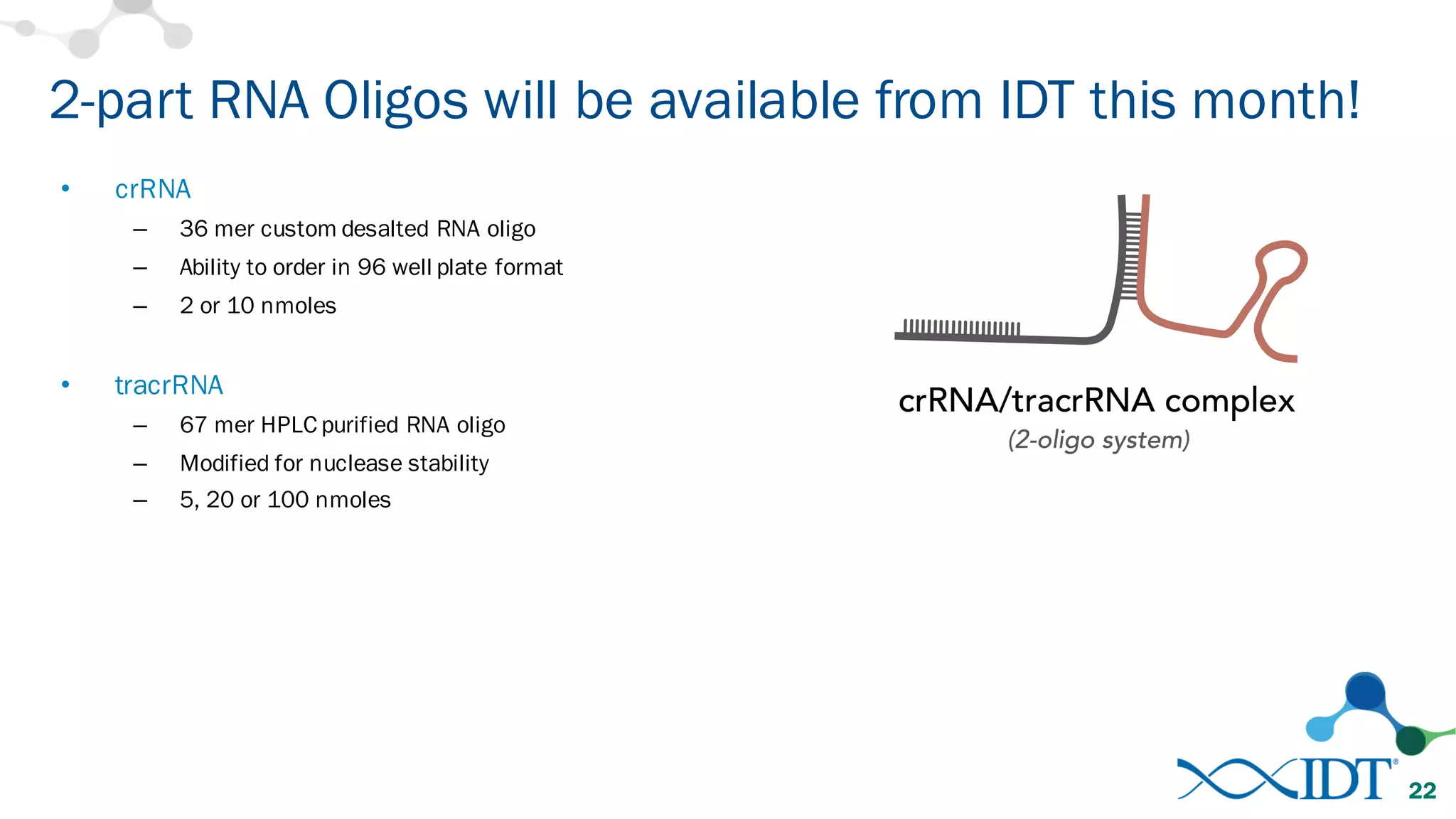
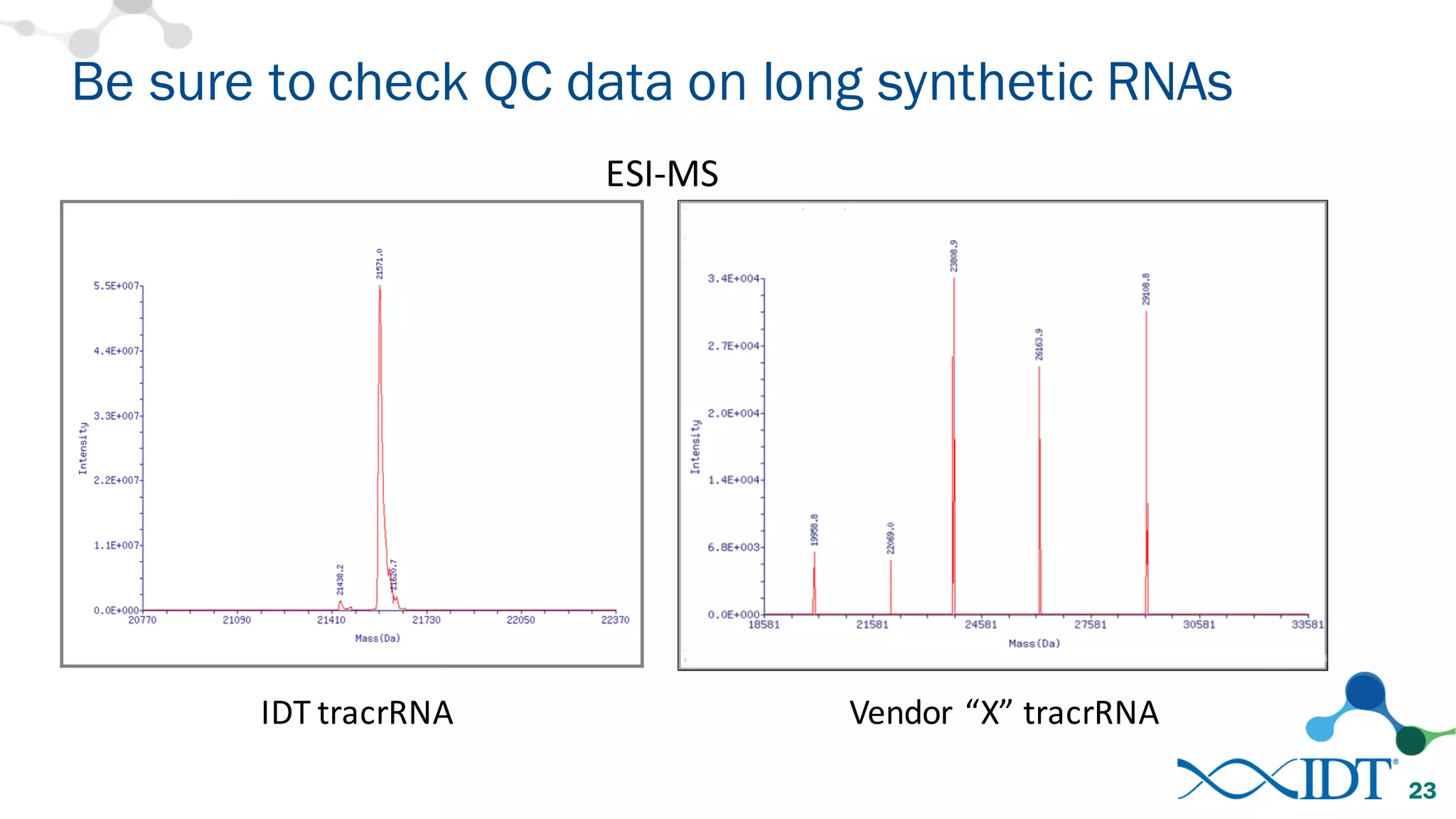
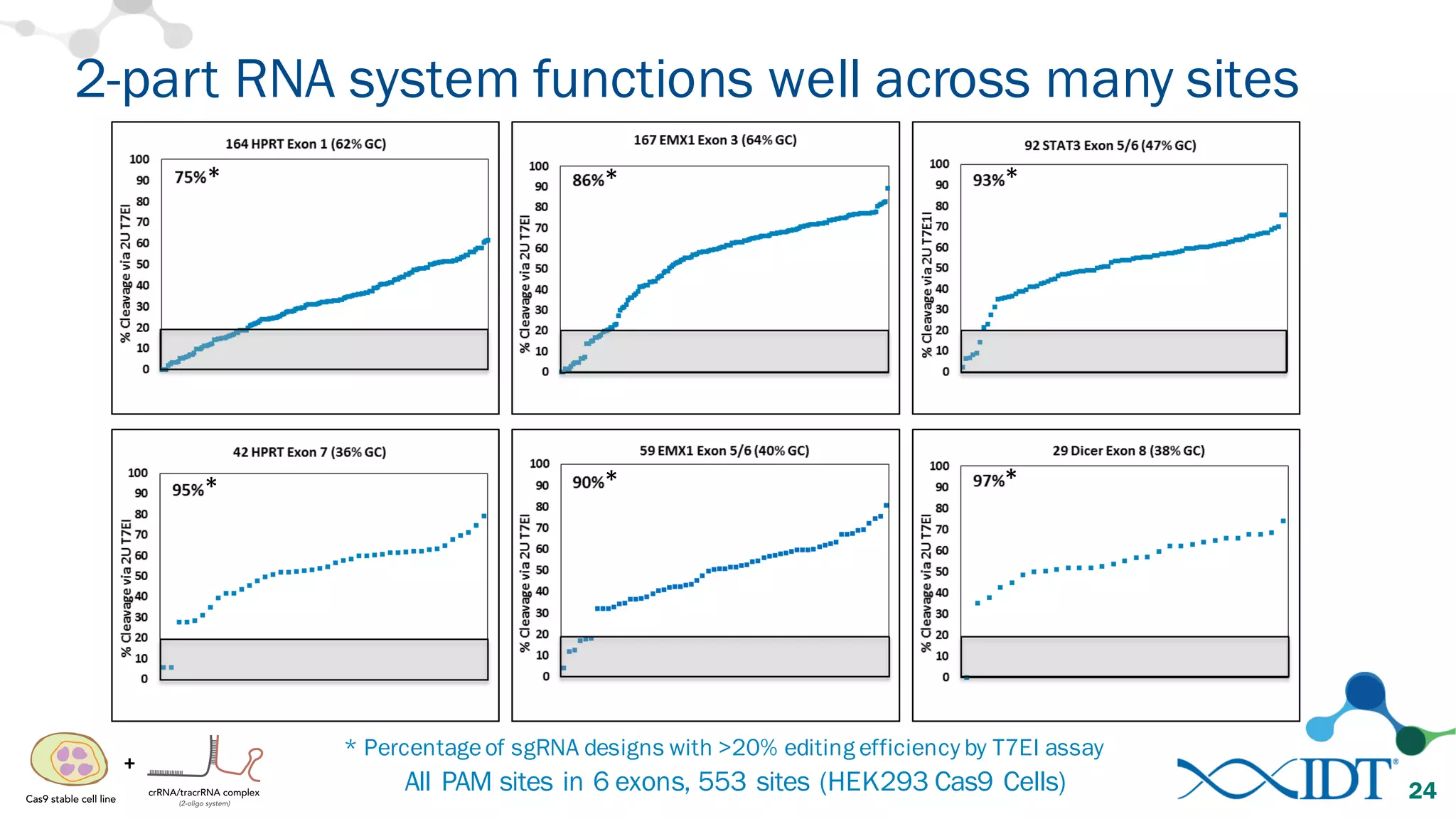
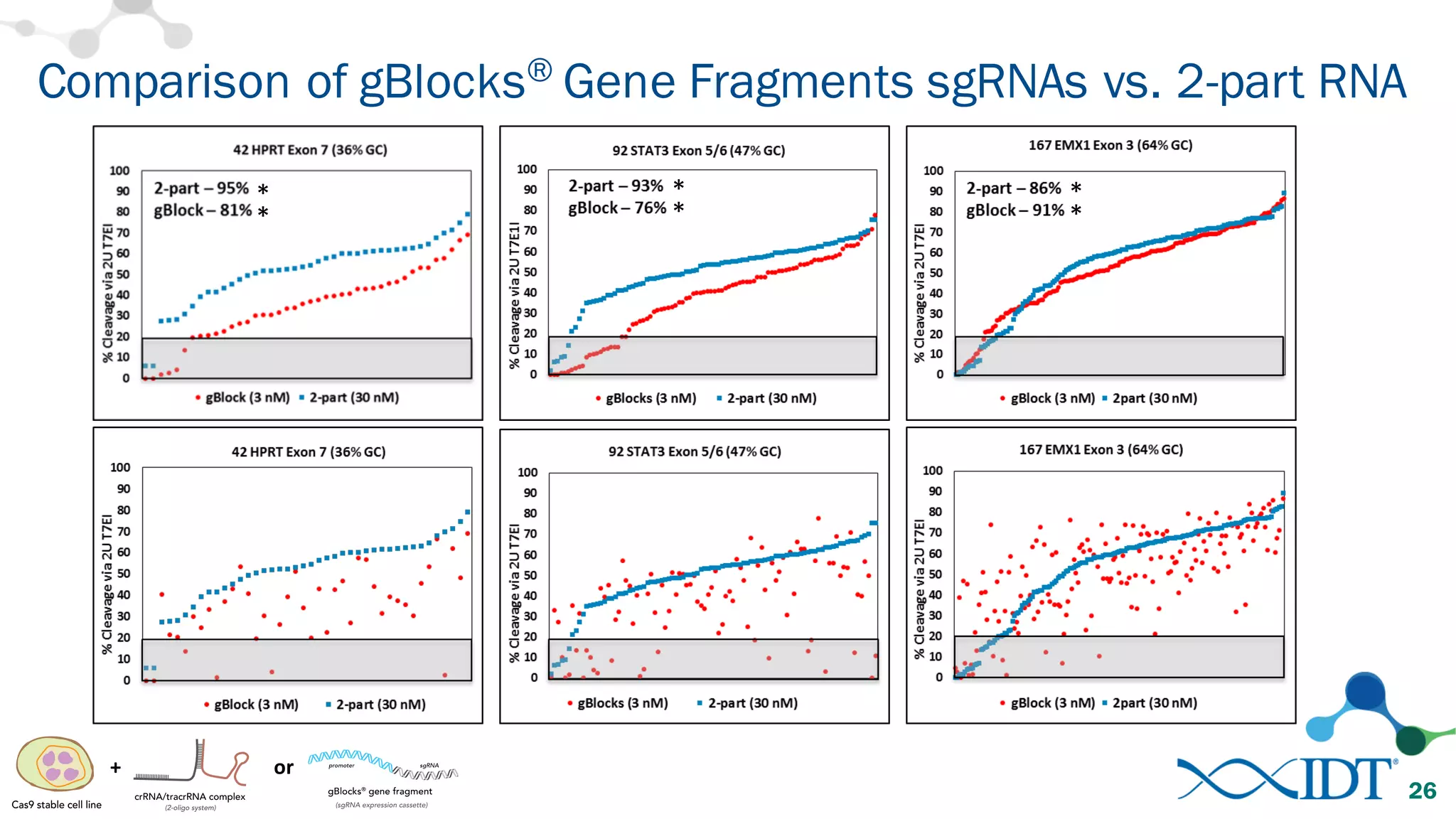
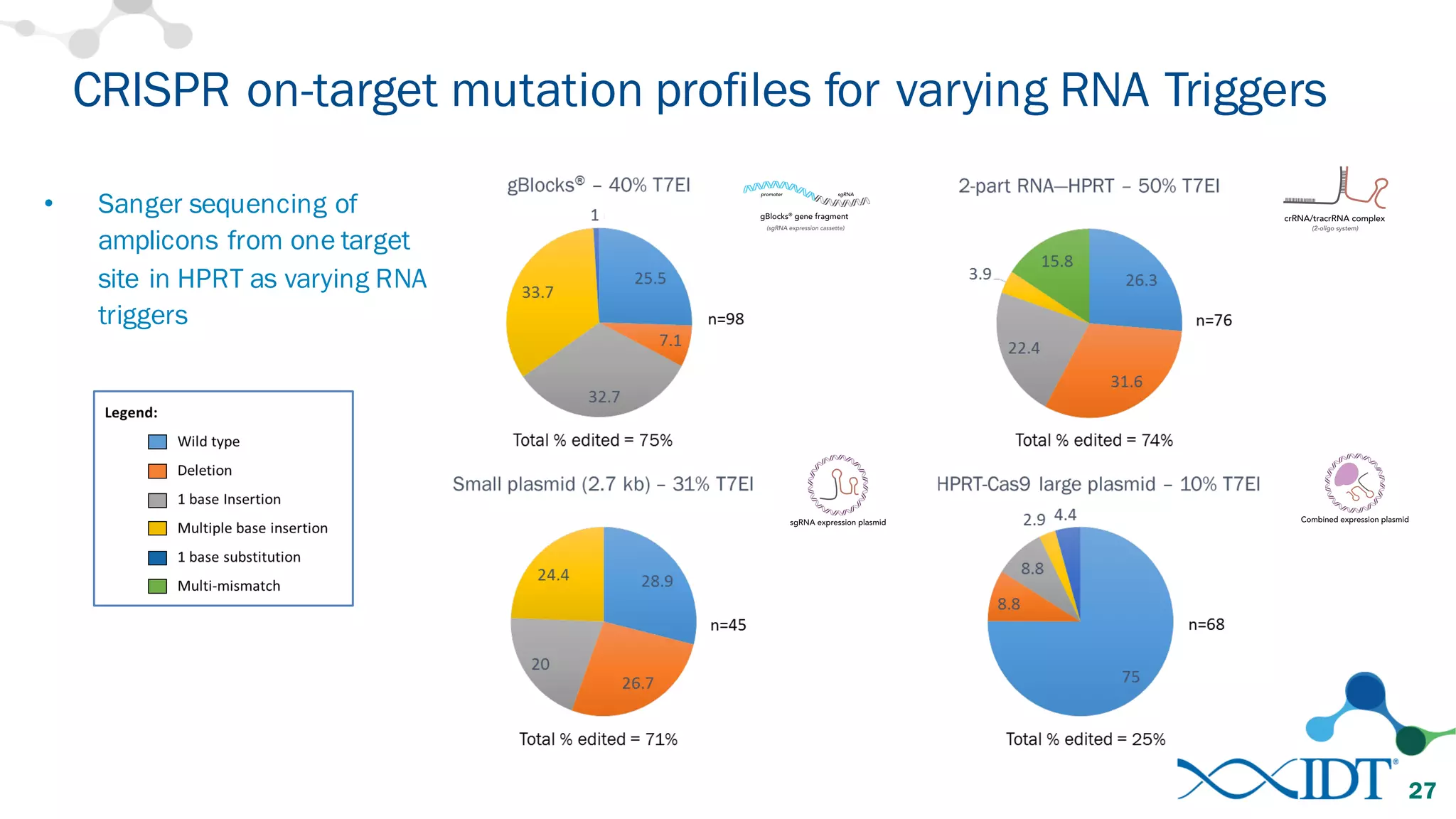
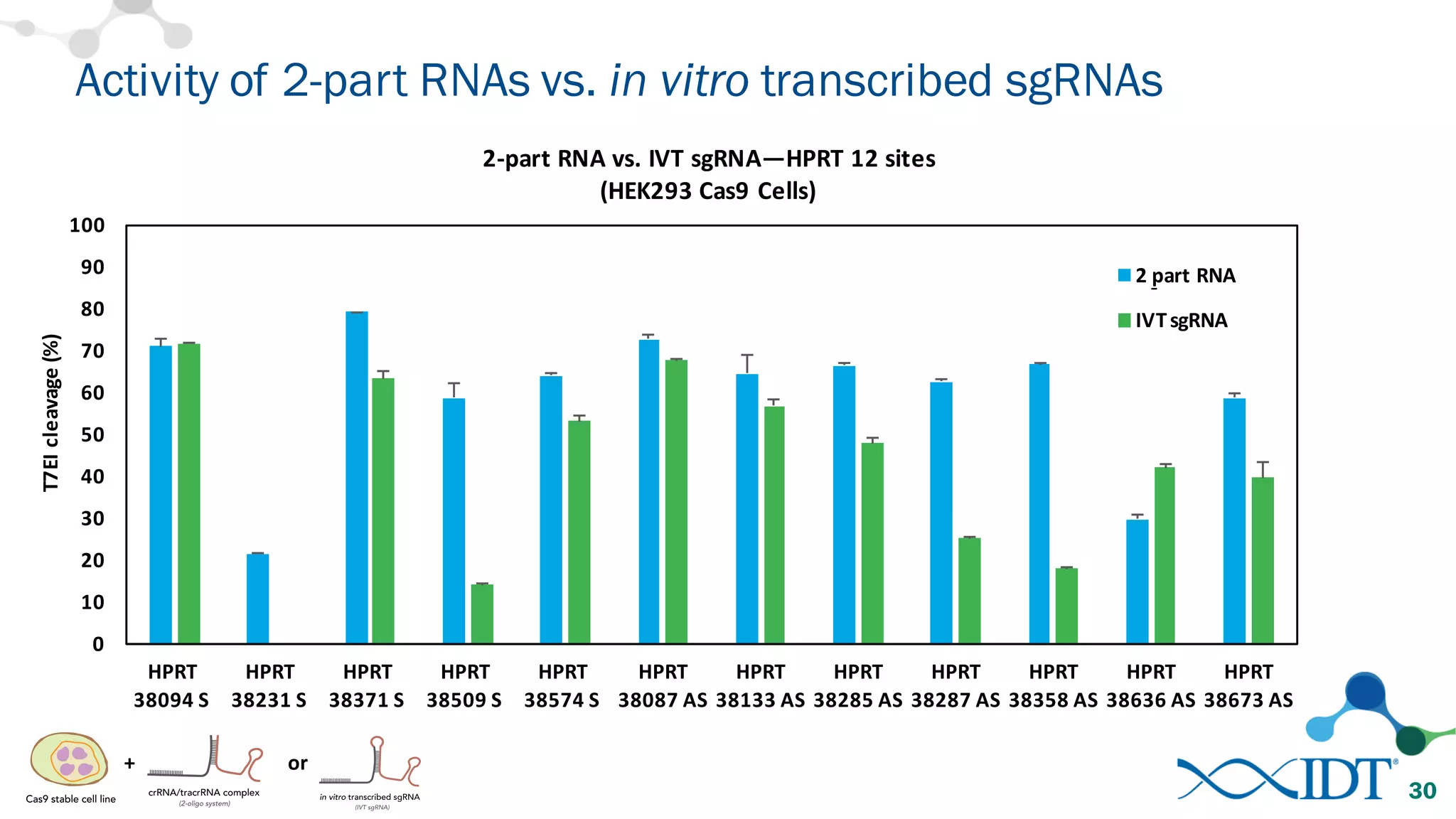
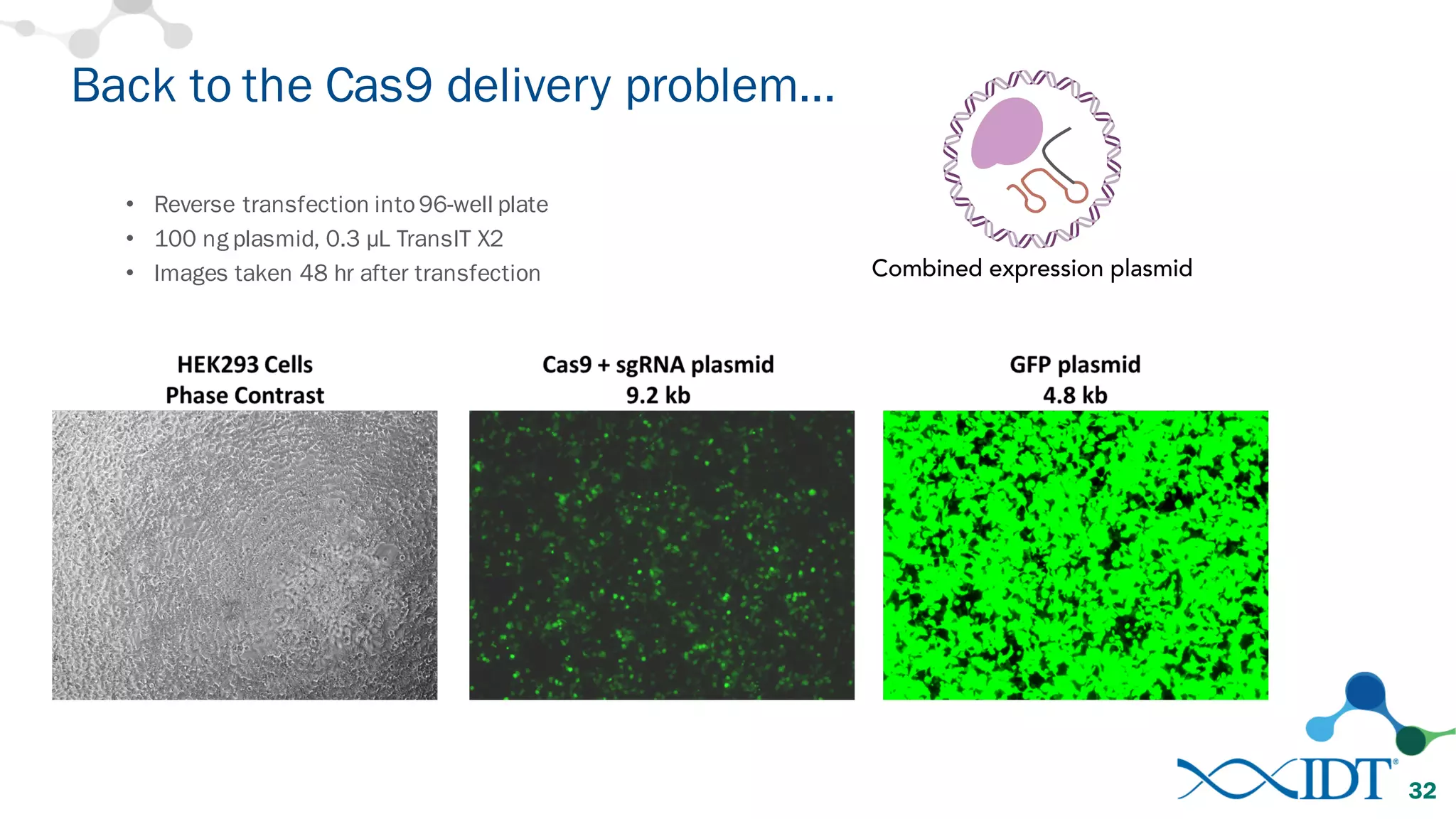
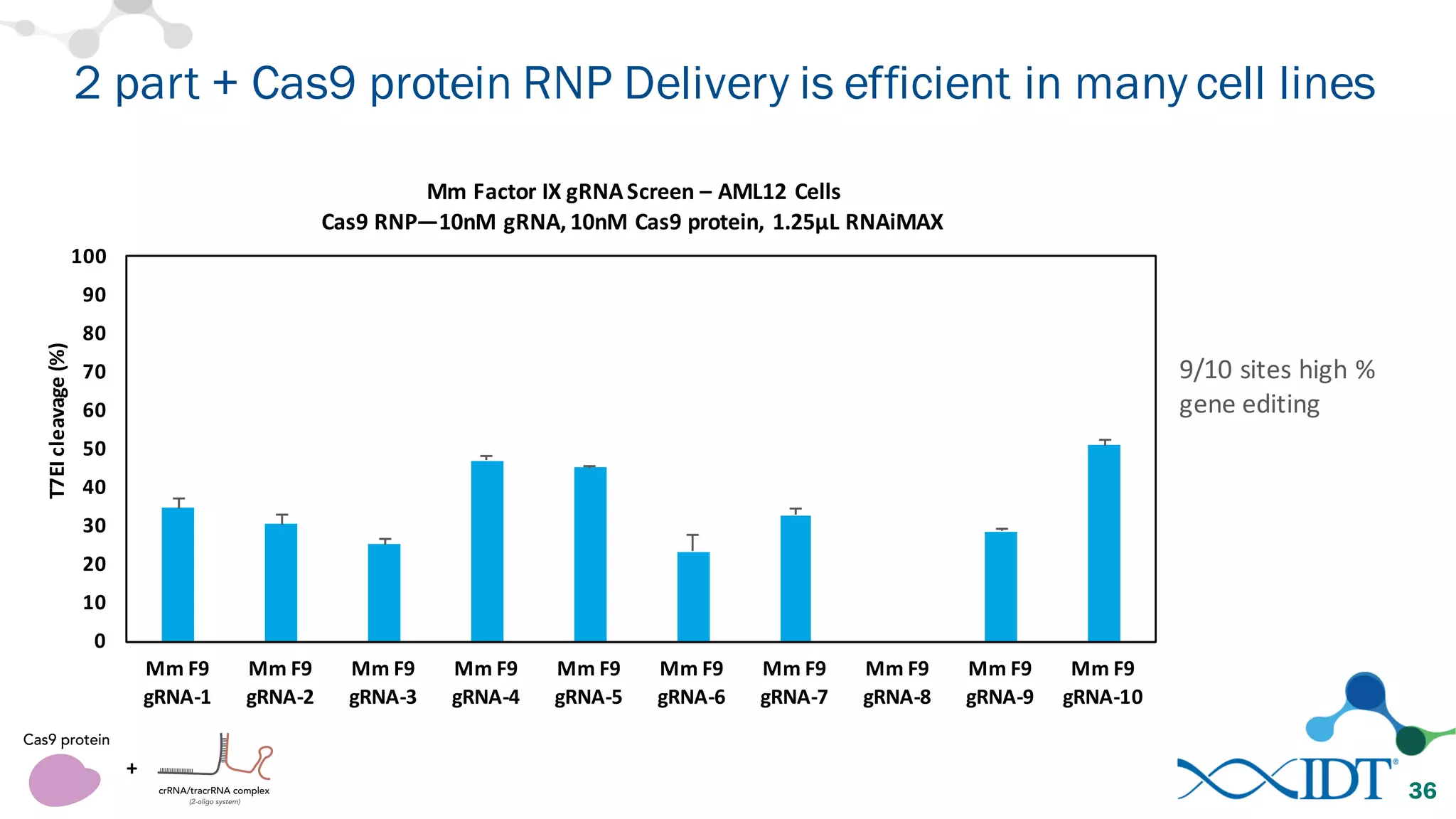
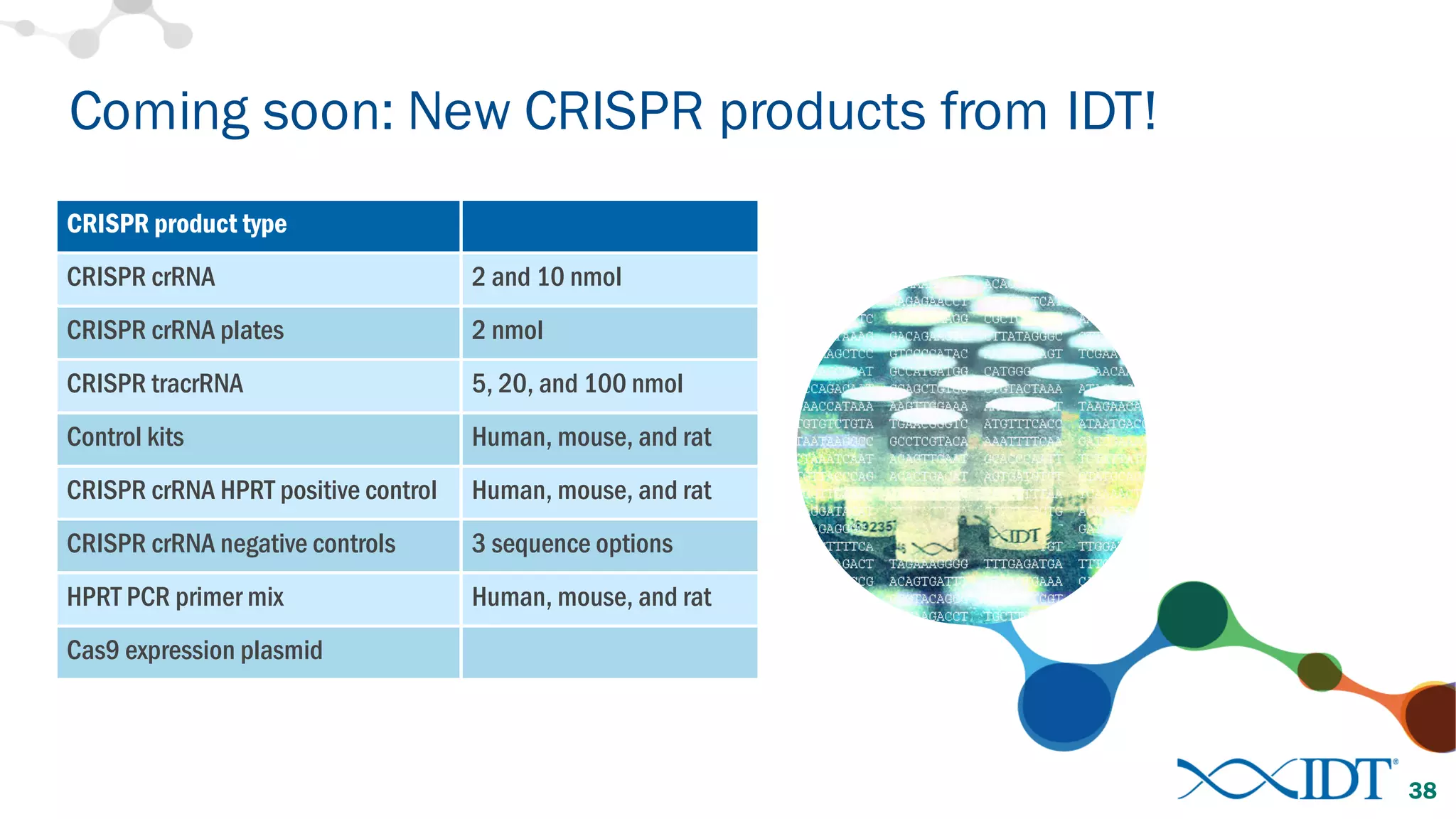
The document discusses advancements in CRISPR/Cas9 genome editing, focusing on solutions for delivering Cas9 and sgRNA, optimizing editing efficiency, and improving methods to reduce off-target effects. It highlights various experimental results and methods for synthesizing gBlocks gene fragments for sgRNA expression, examining the performance of different RNA configurations. Key findings indicate that the use of two-part RNA oligos enhances editing efficacy and reduces immunogenic responses compared to in vitro transcribed sgRNAs.