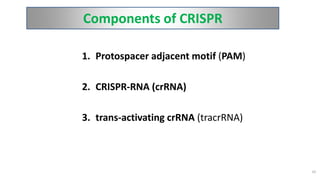
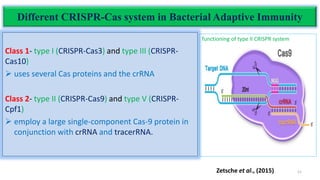
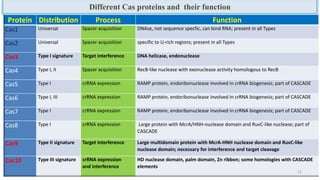
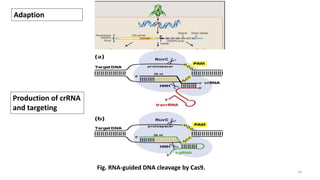
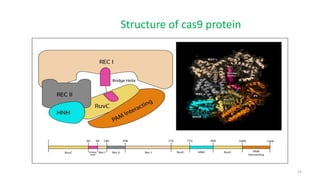
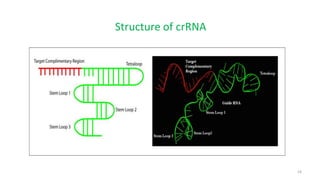
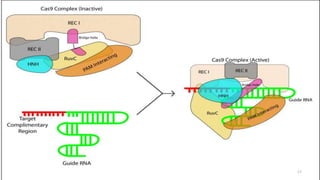
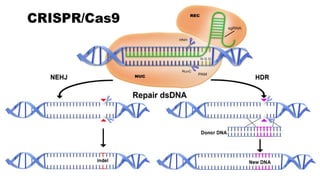
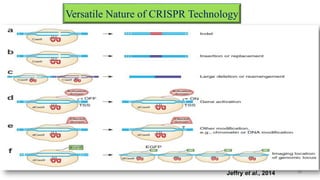
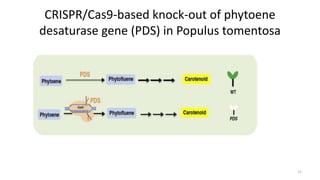
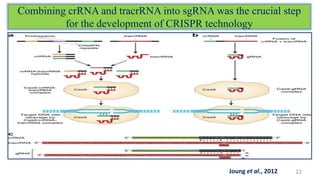
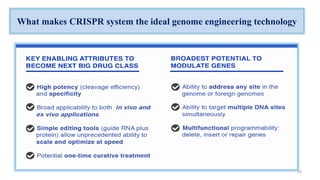
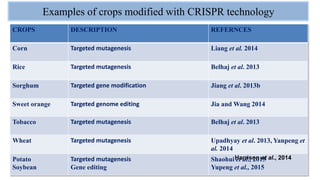
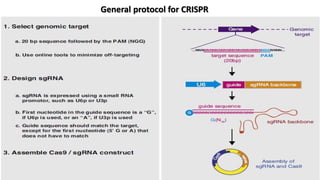
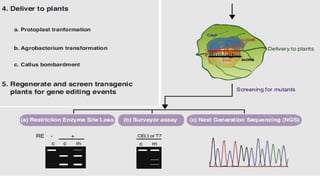
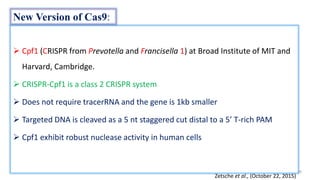
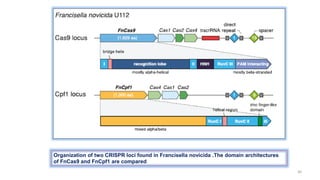
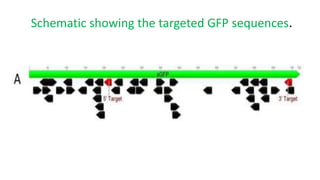
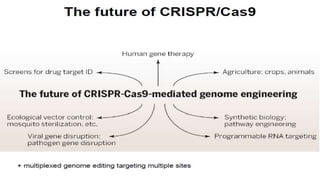
The document discusses CRISPR-Cas9 as a revolutionary genome editing tool, detailing its history, structure, mechanism, applications, and advantages over traditional methods. It highlights its significance in genetic engineering, particularly in crops, allowing for precise modifications that can enhance desirable traits. Additionally, it addresses recent advances and future potential in agricultural applications and biotechnology.