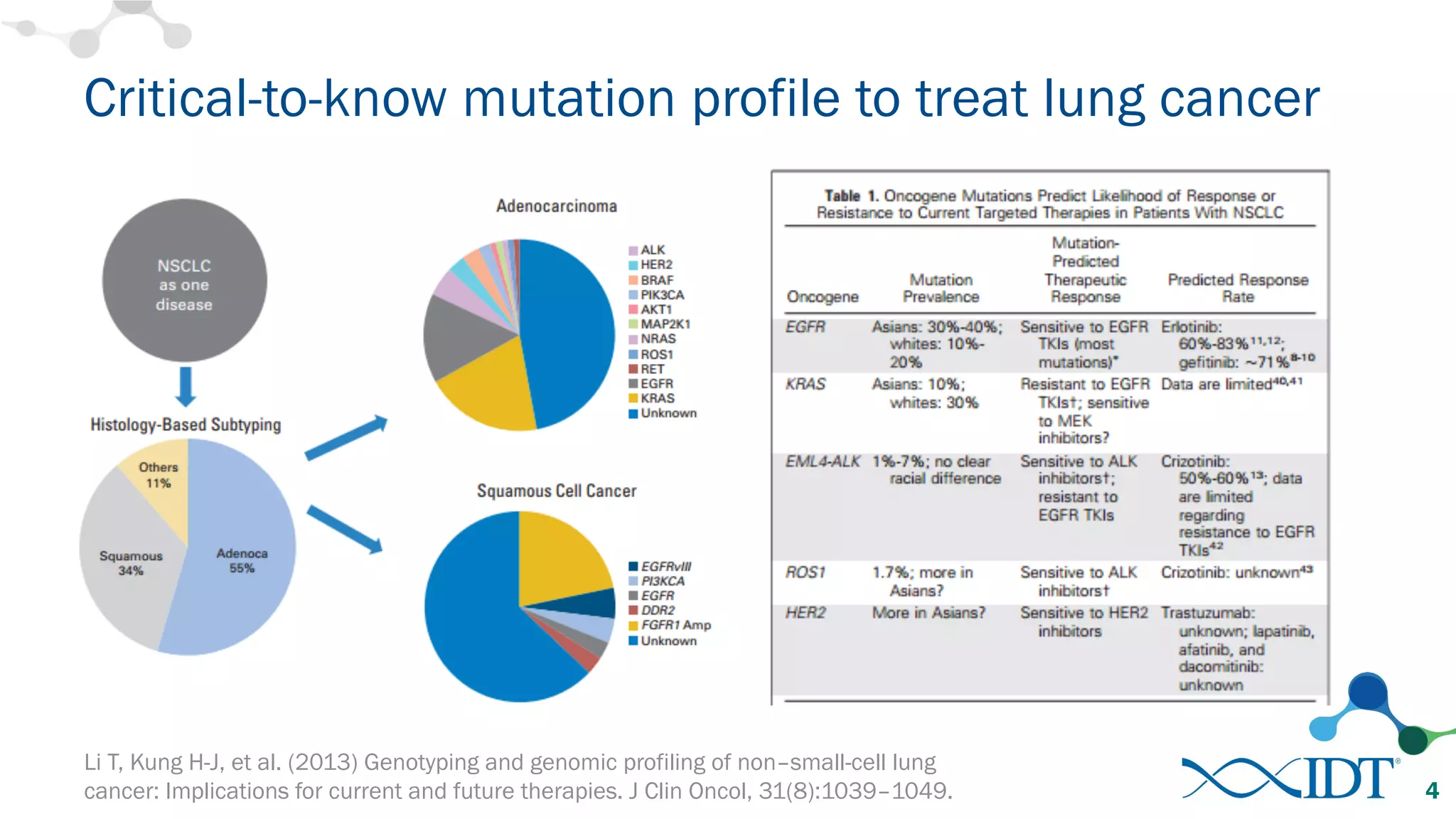
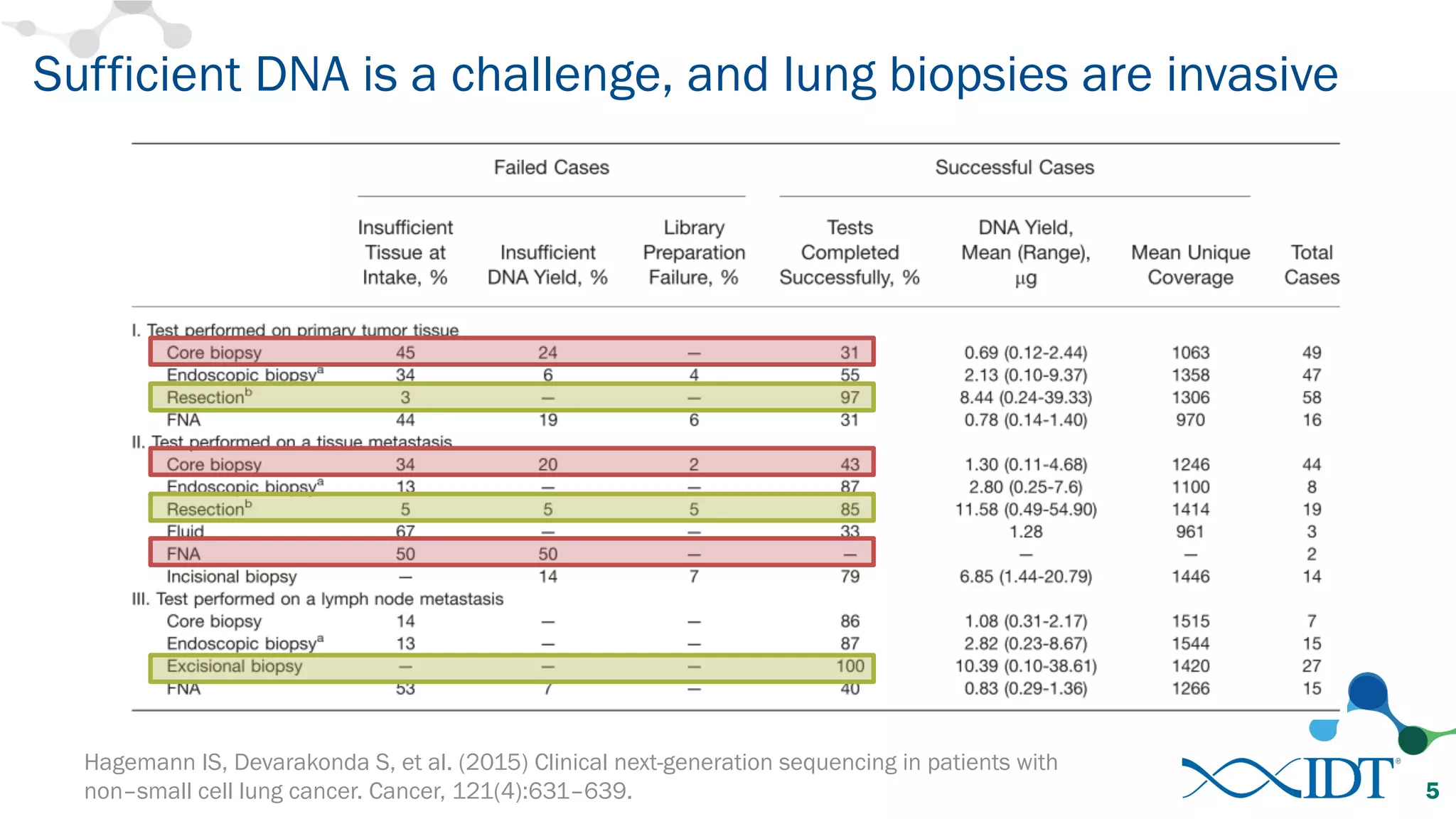
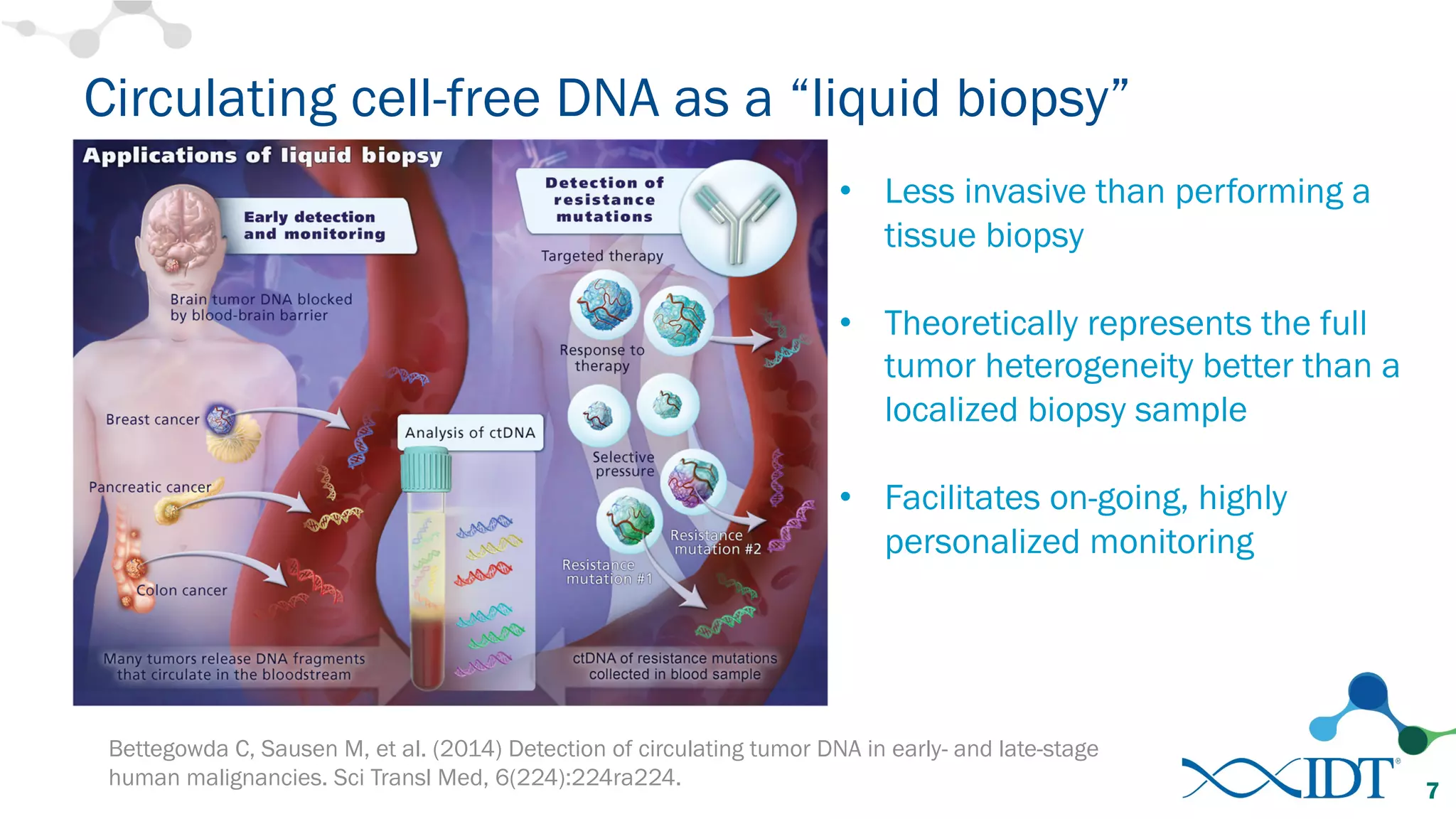
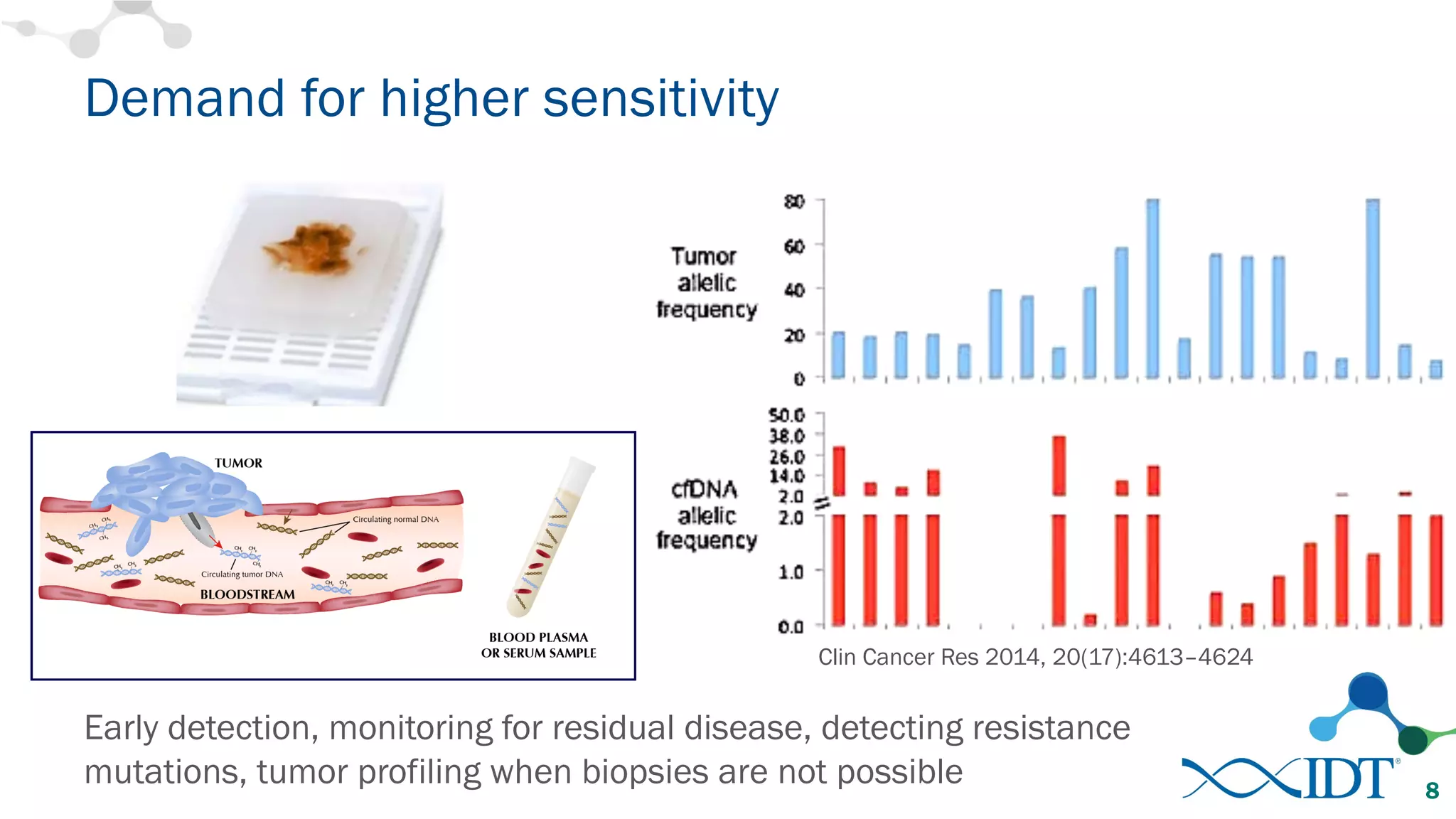
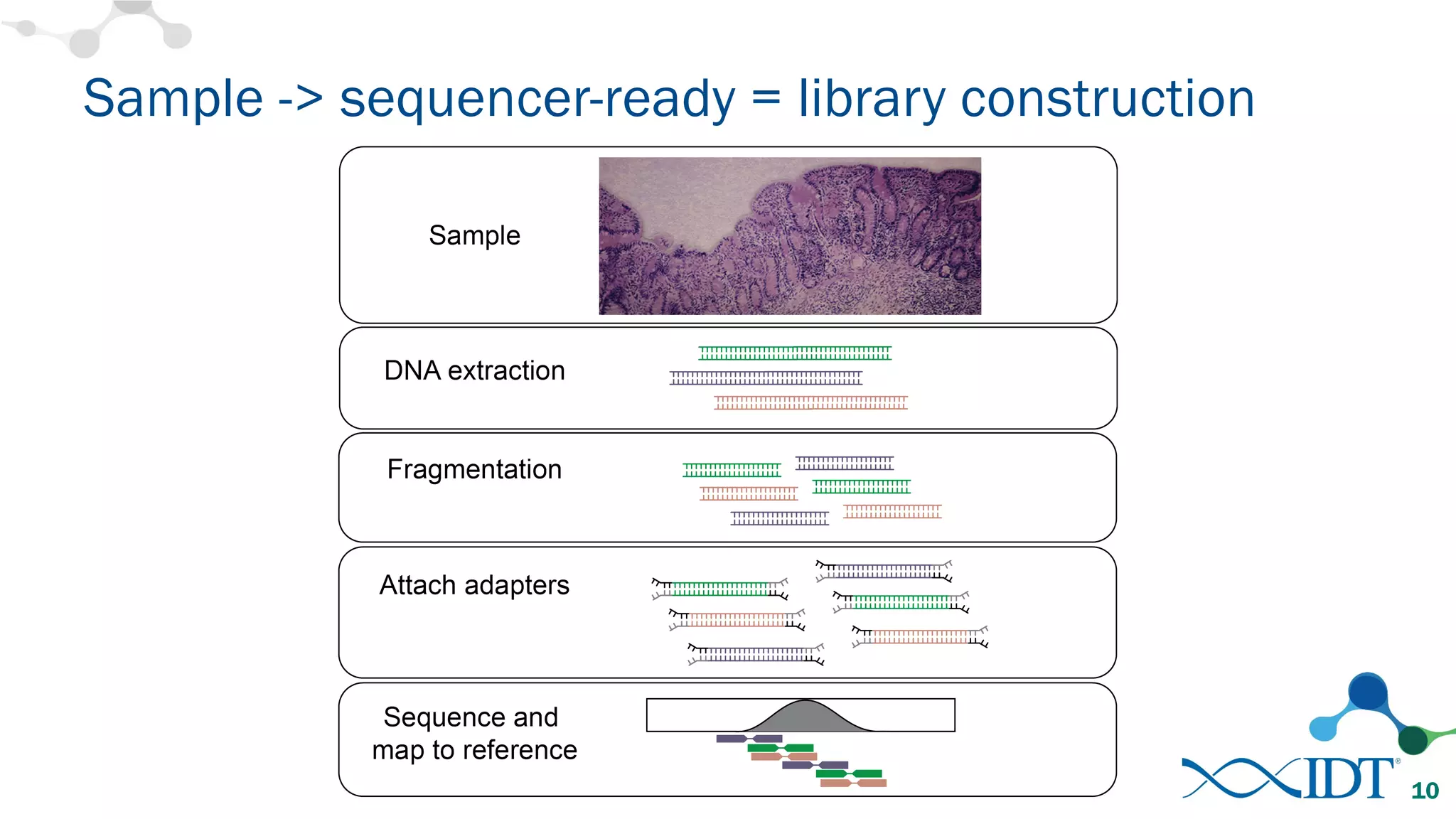
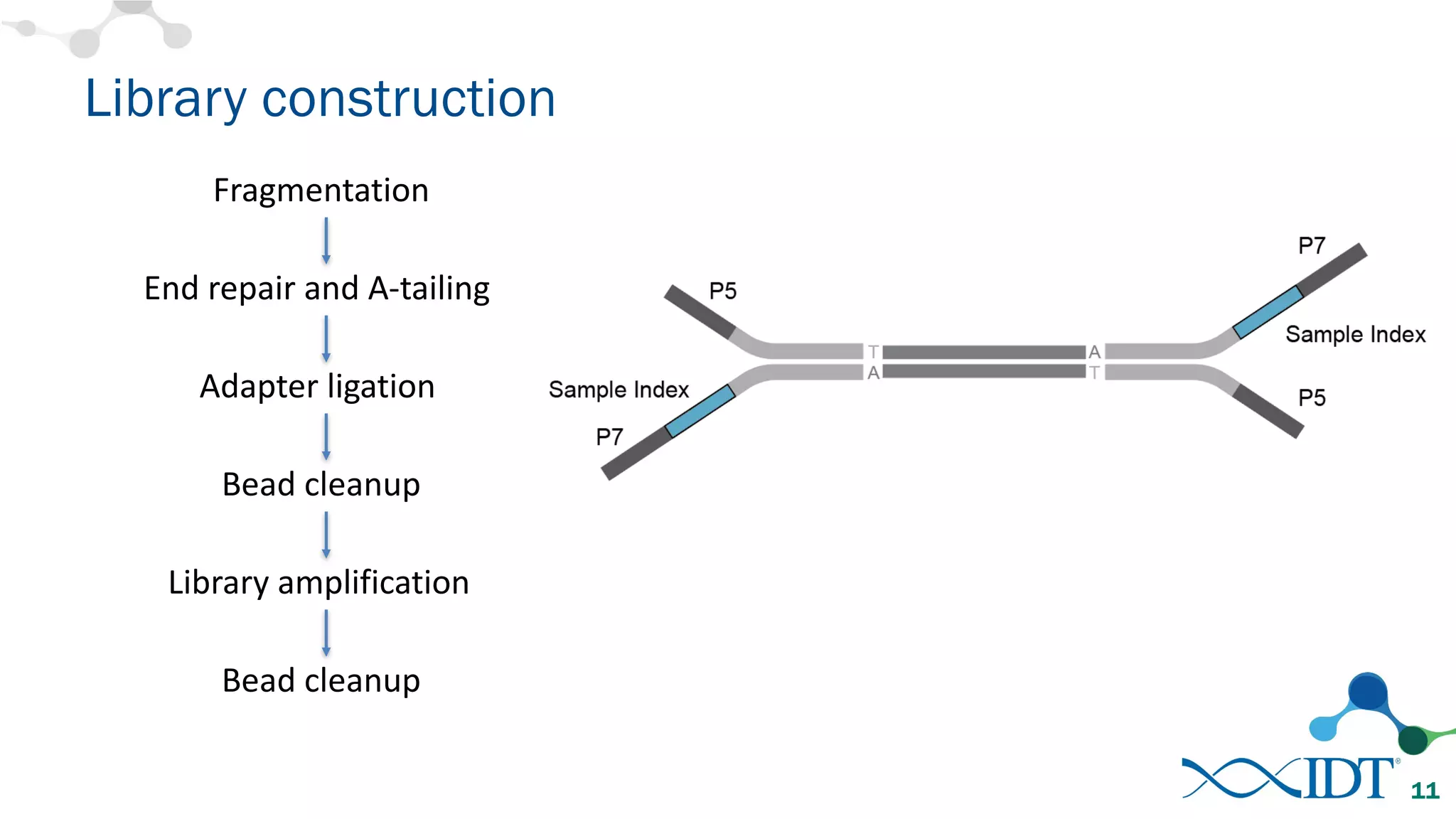
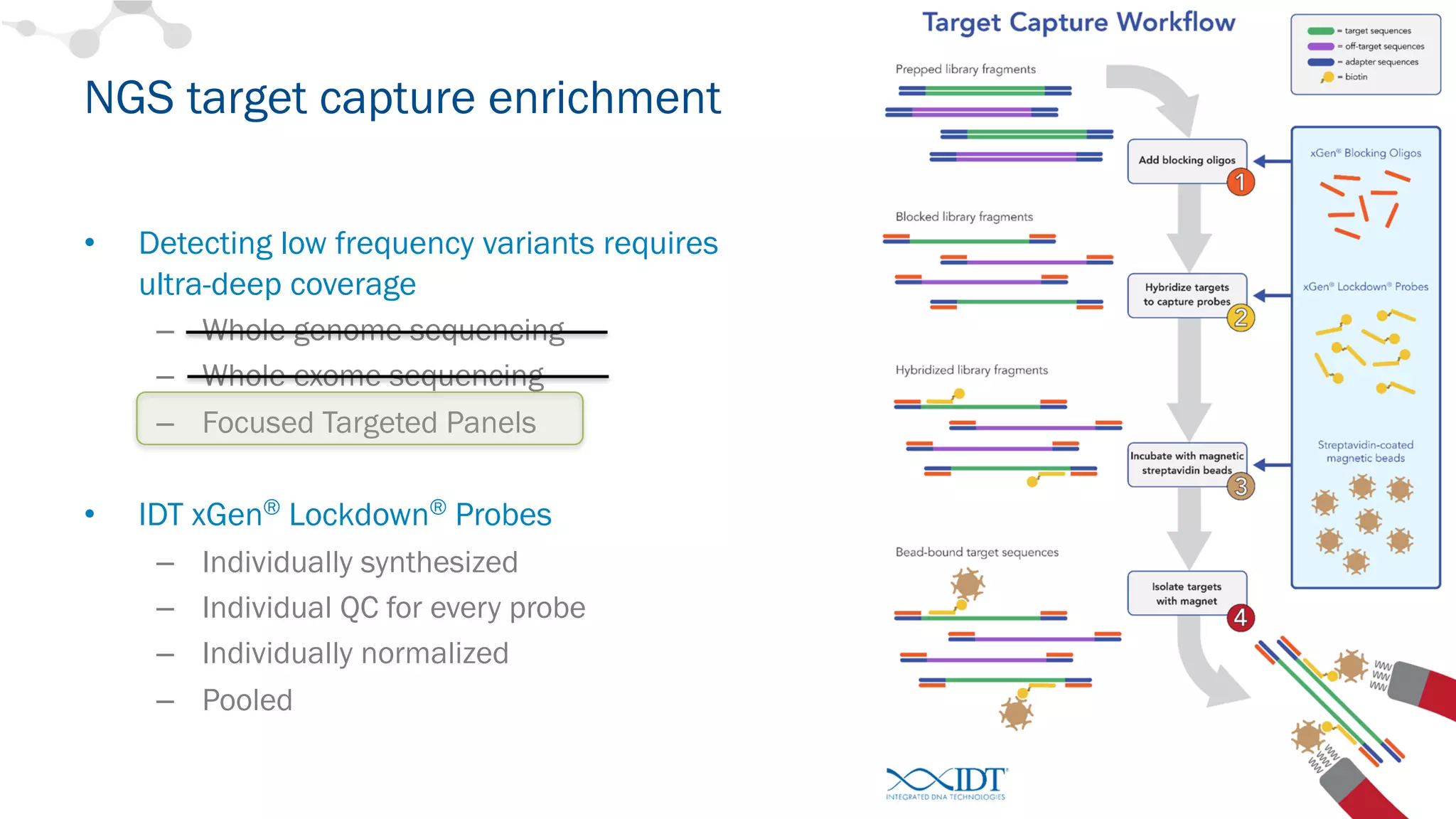
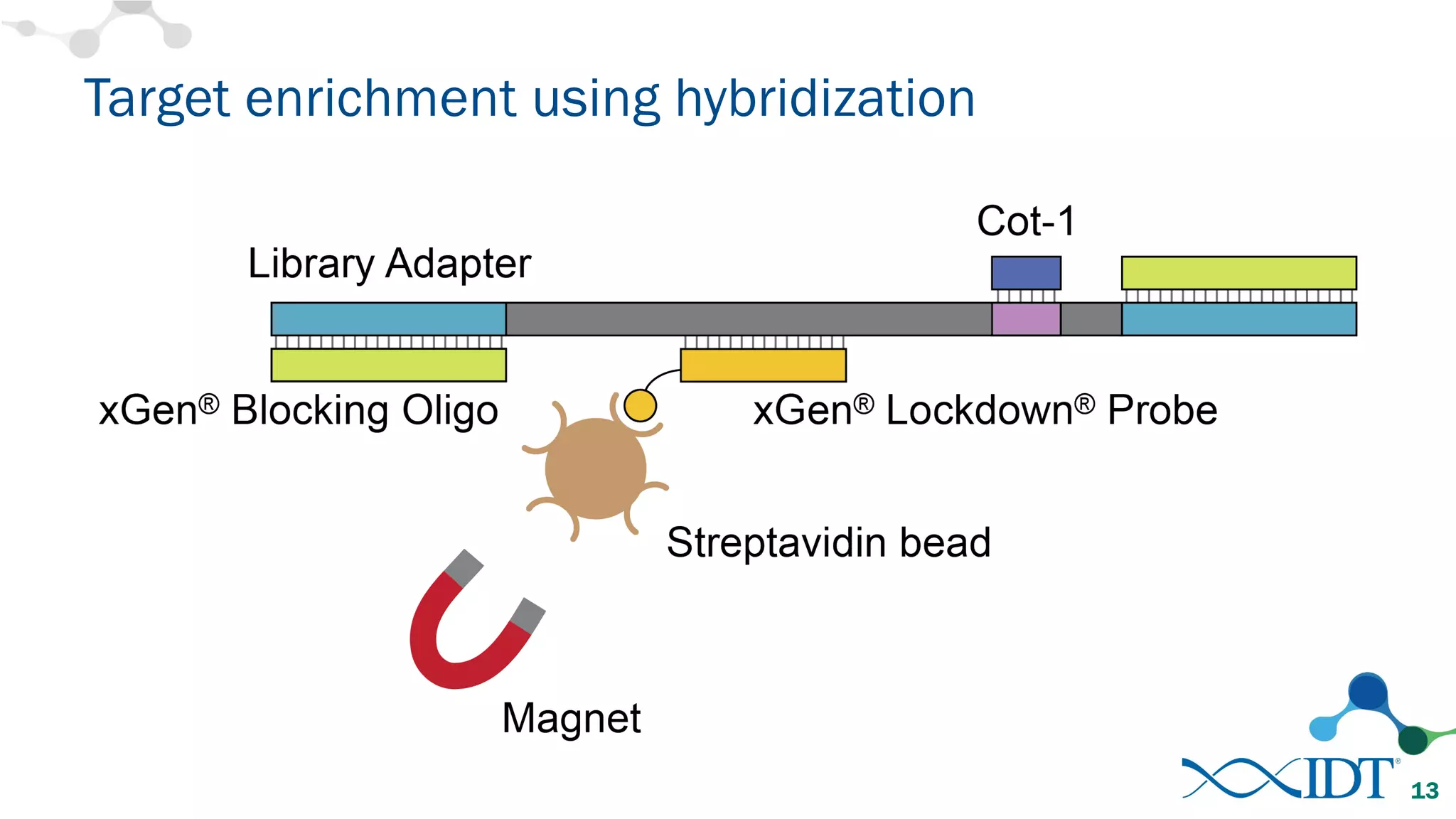
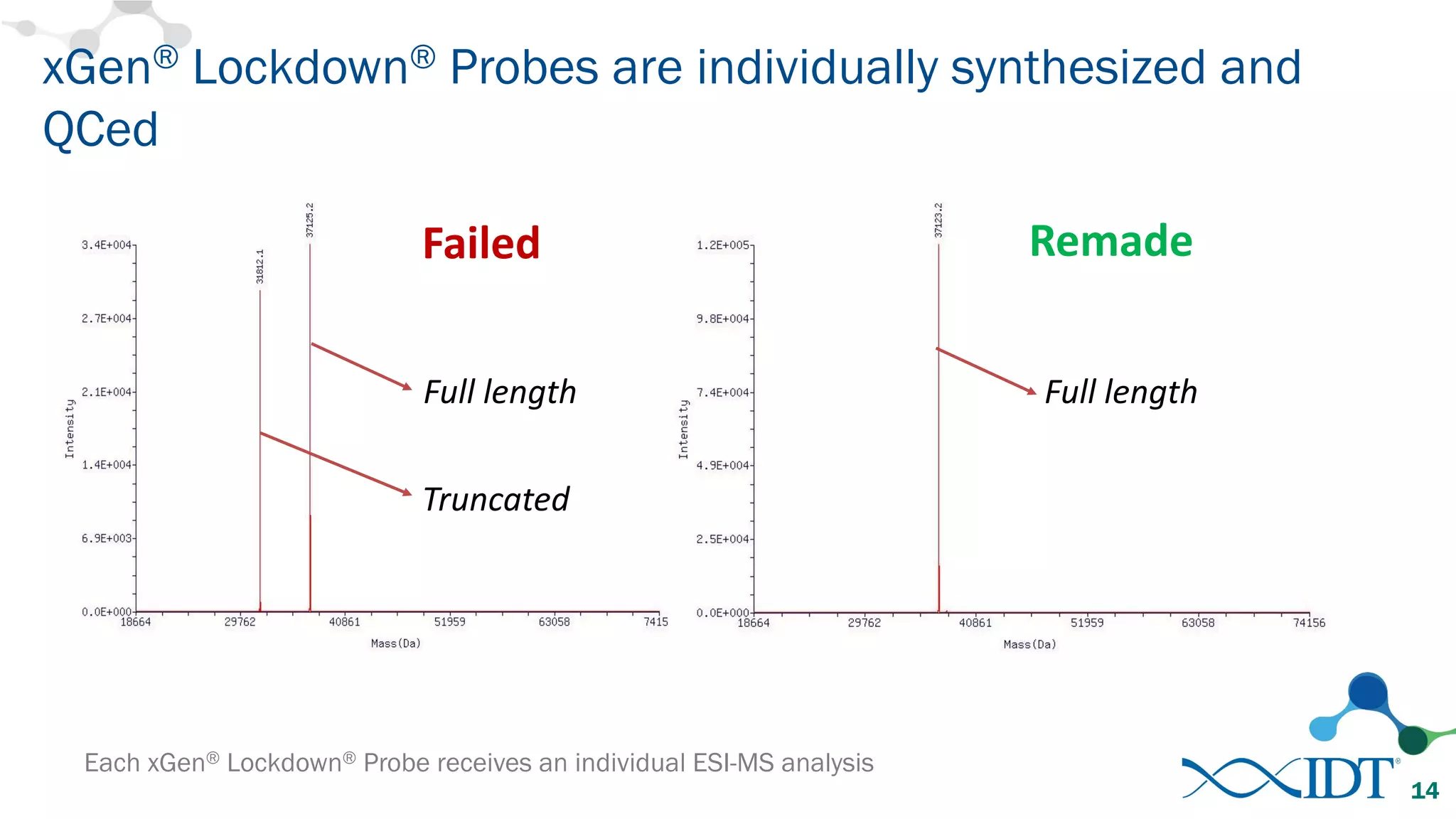
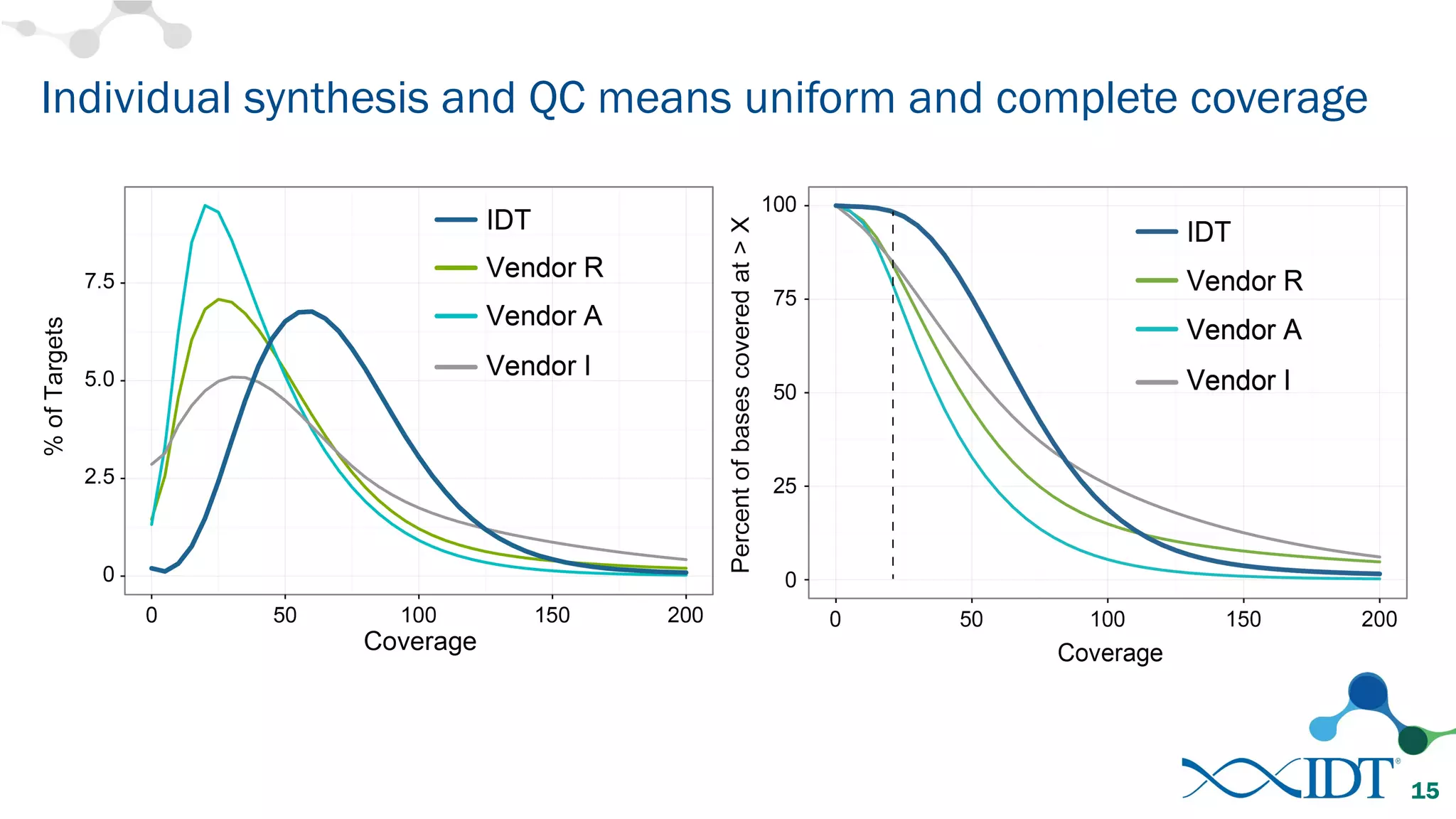
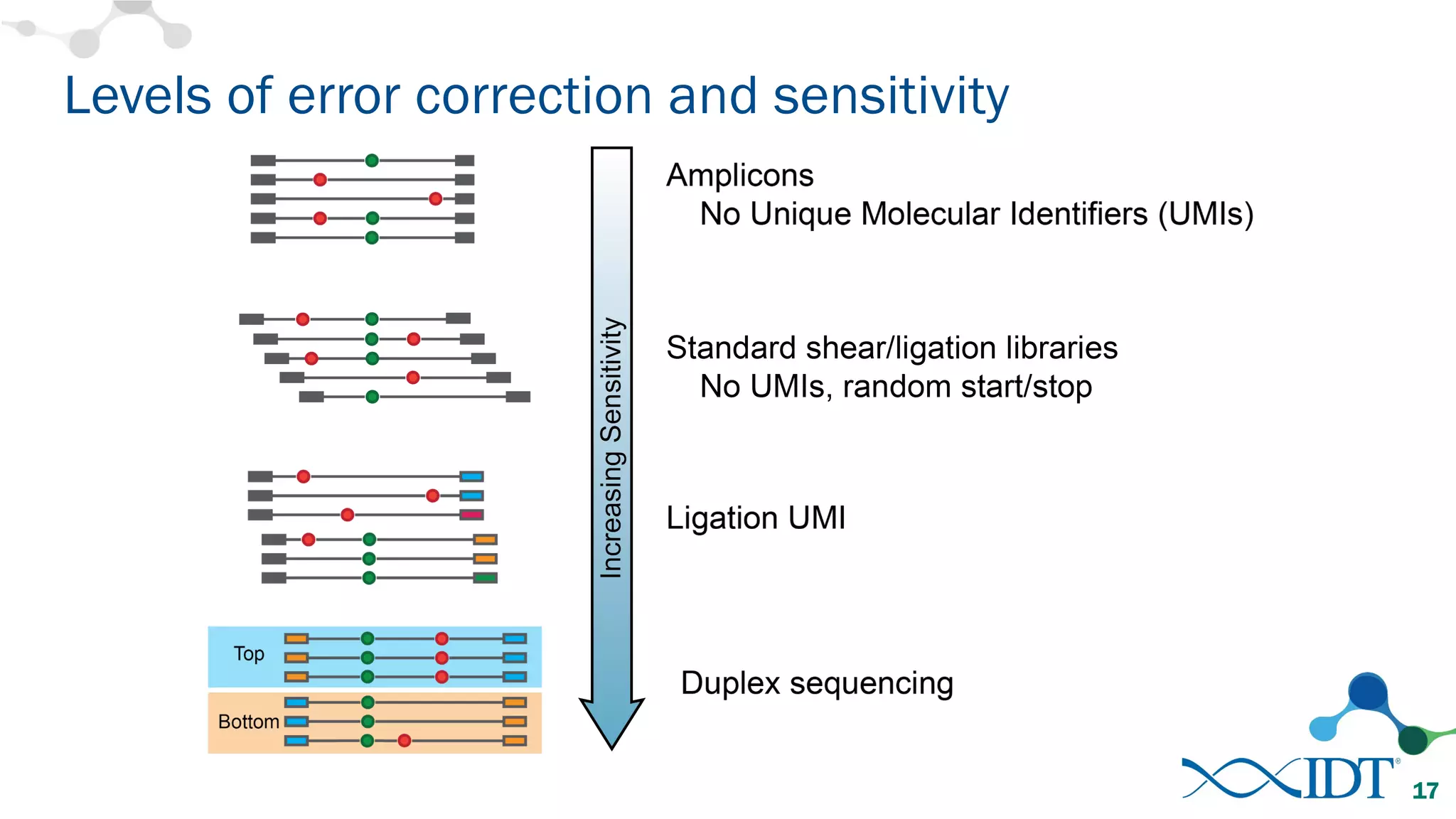
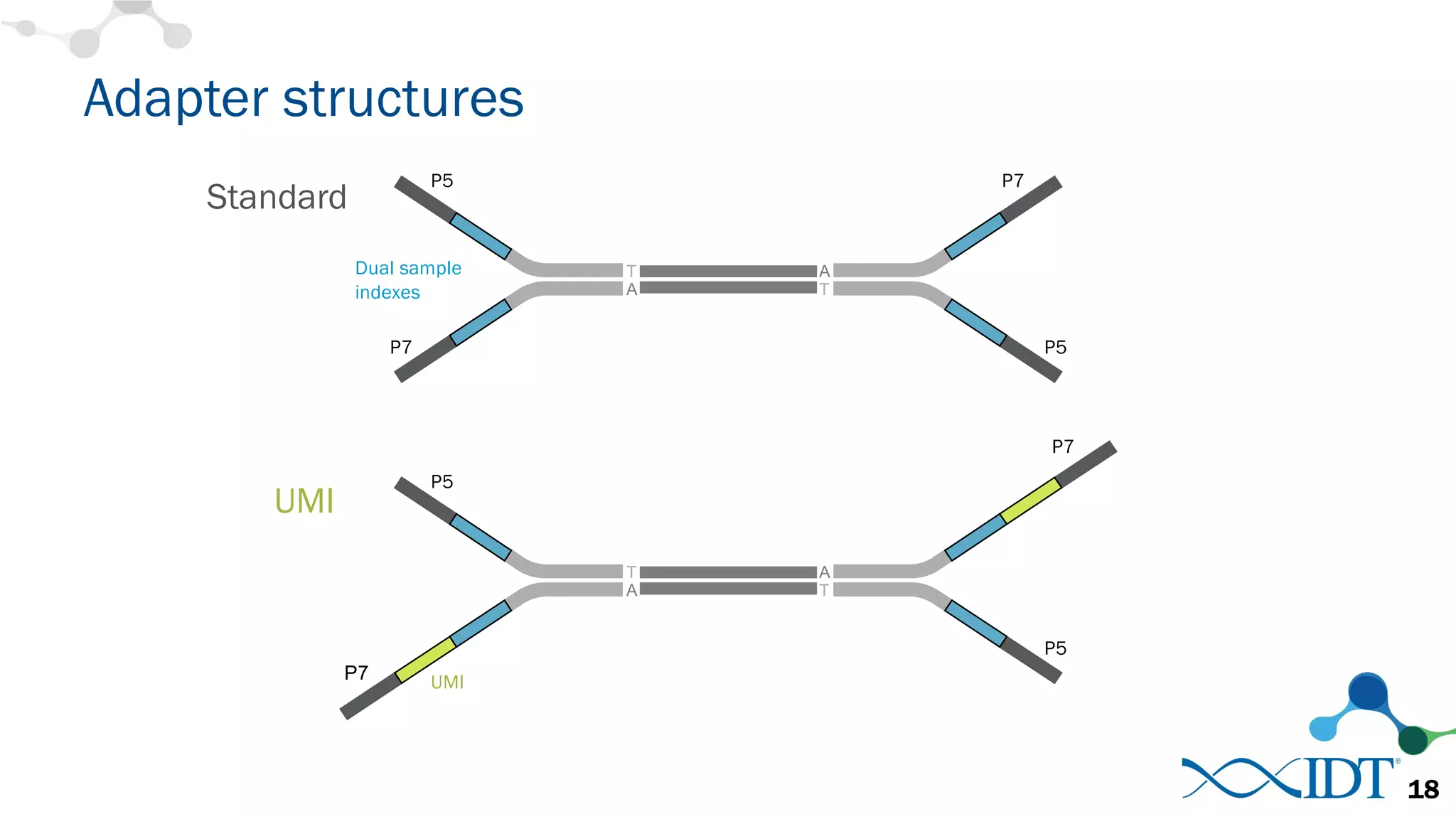
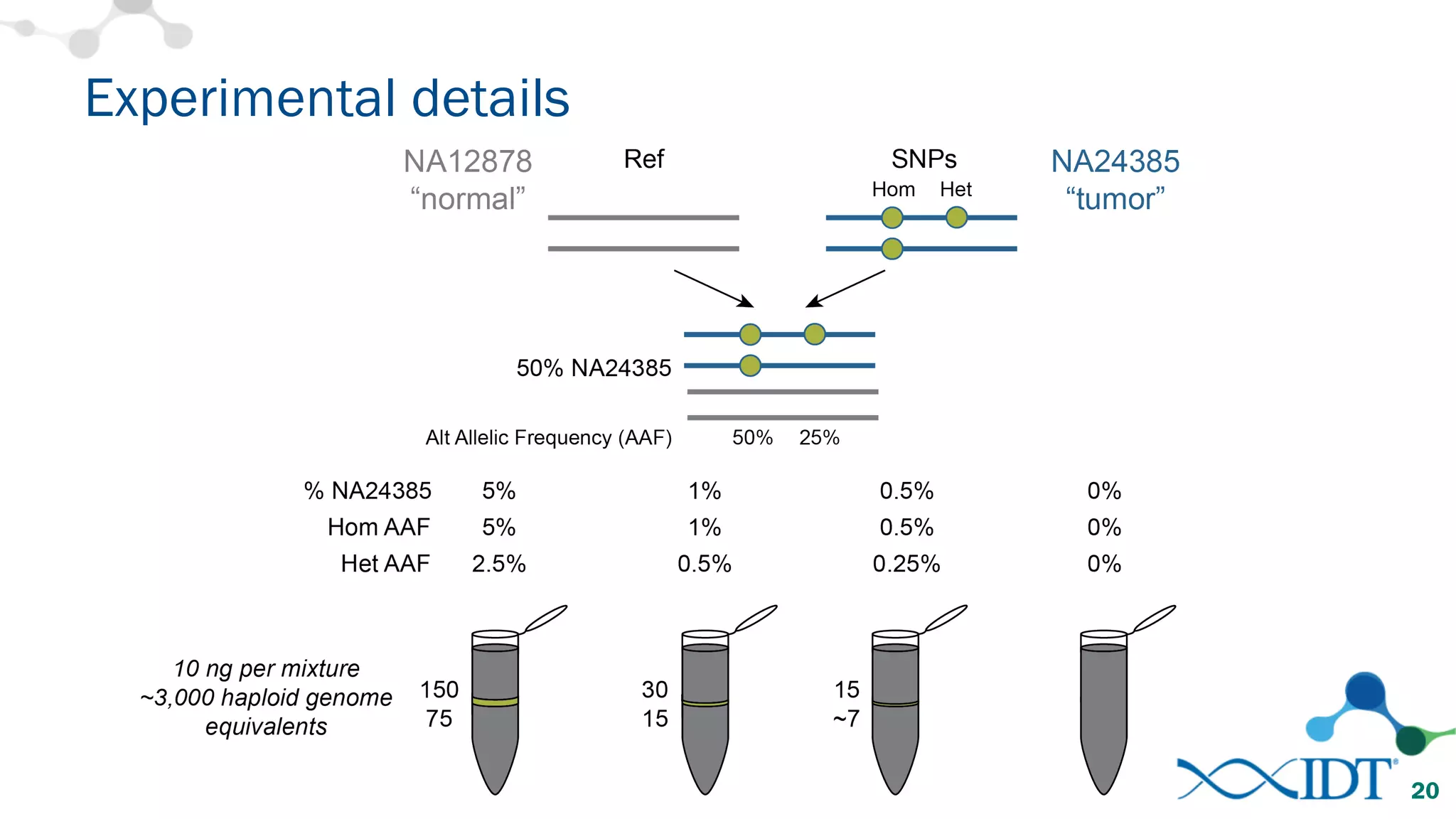
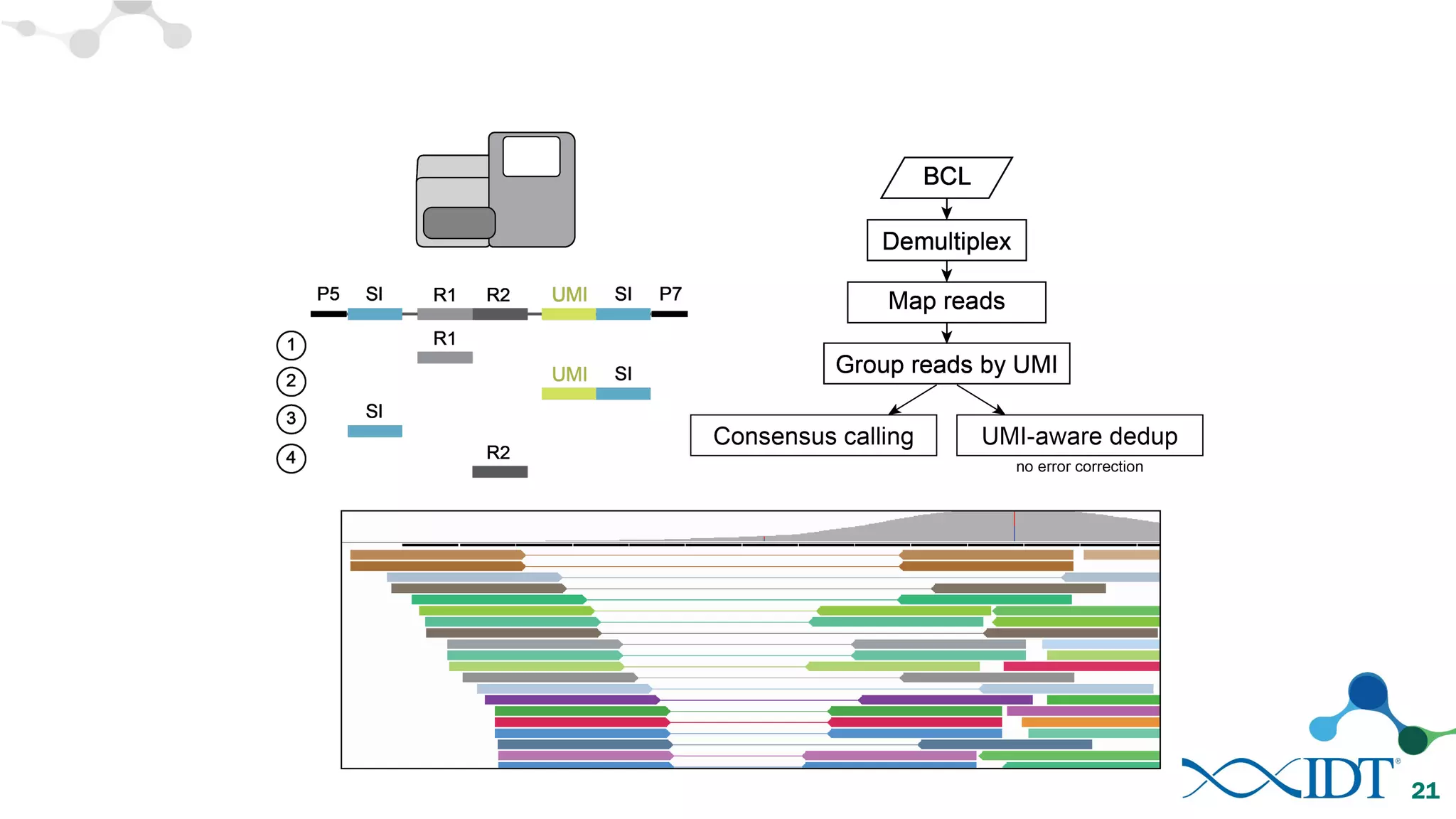
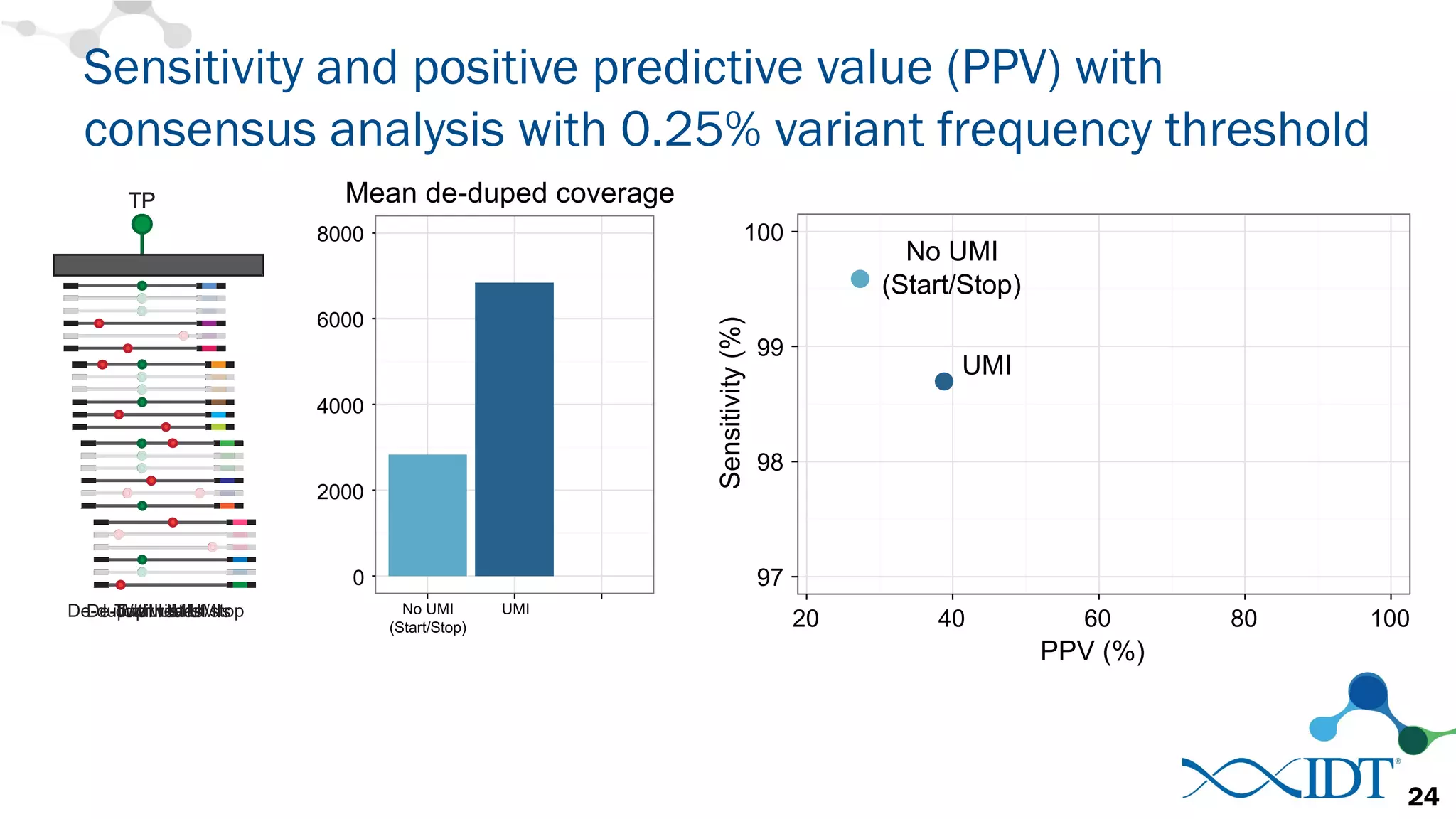
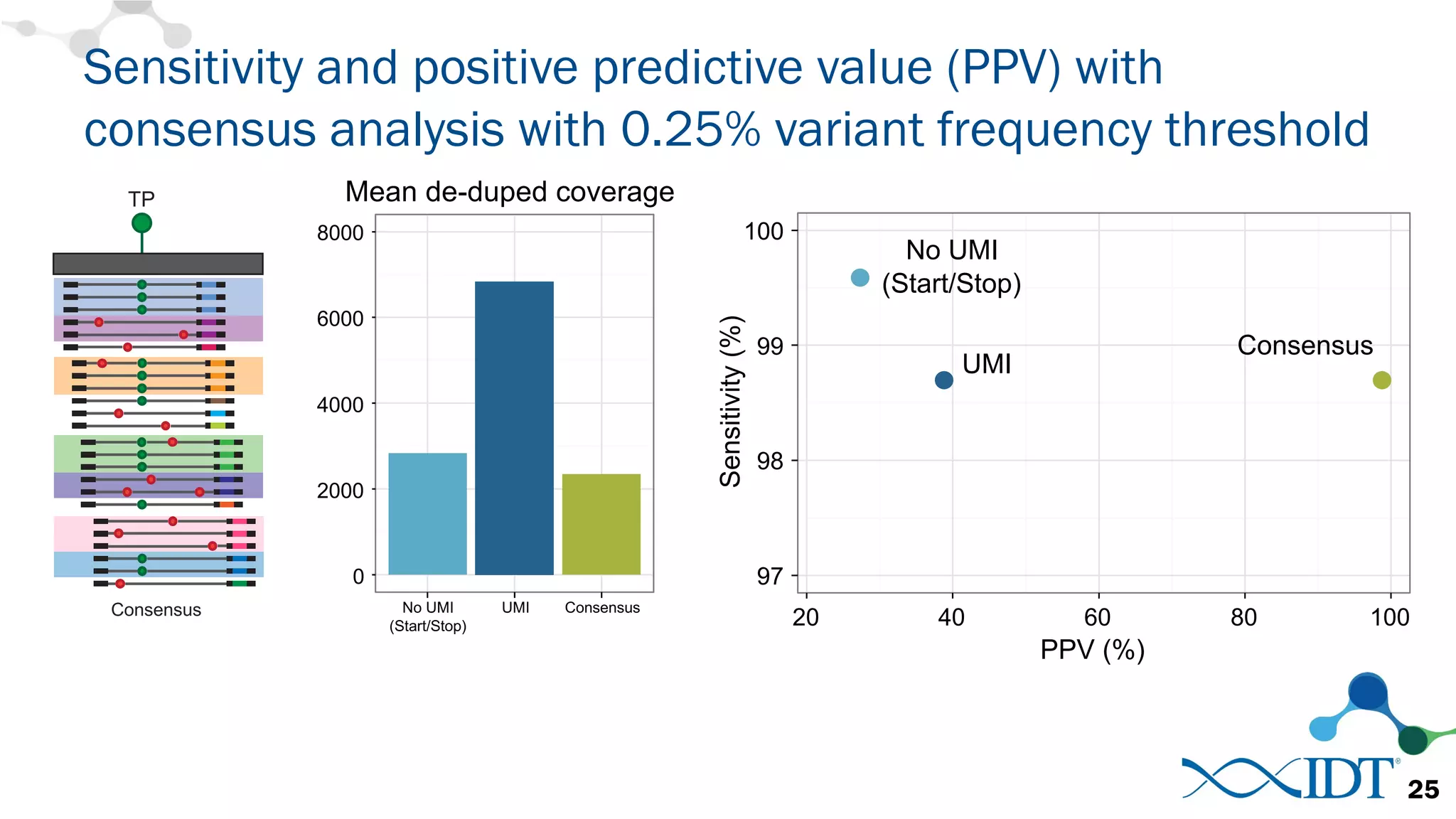
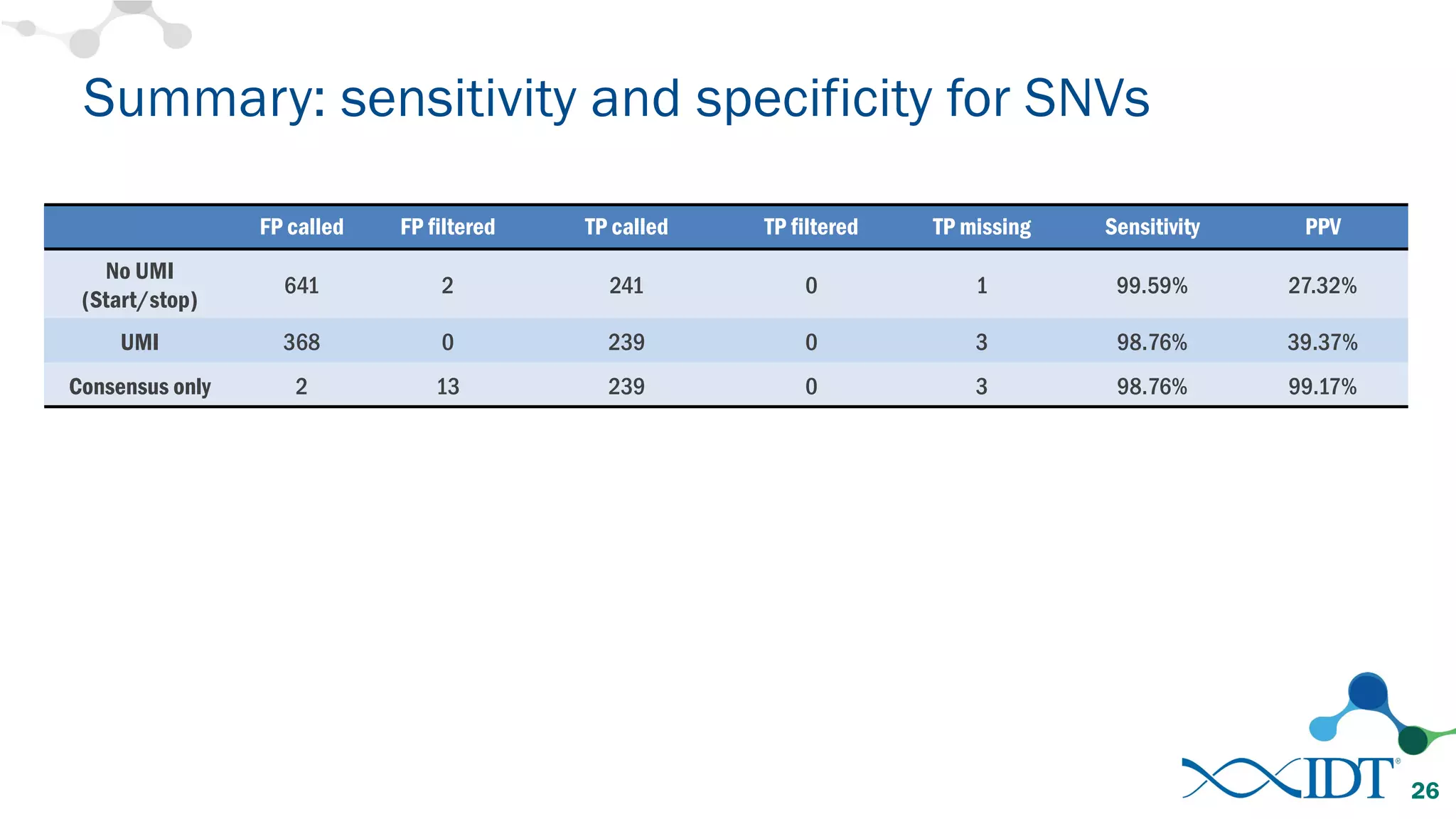
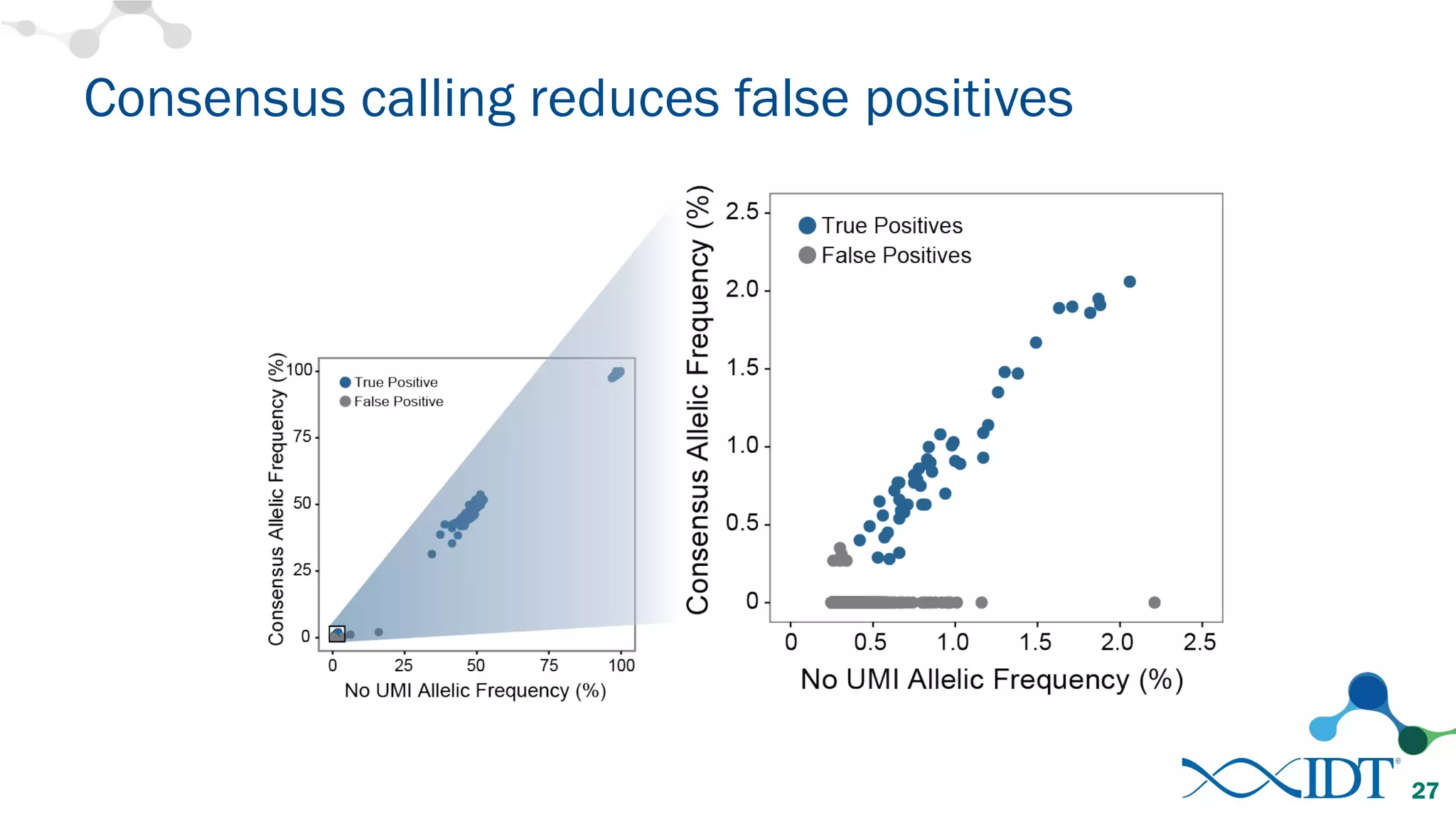
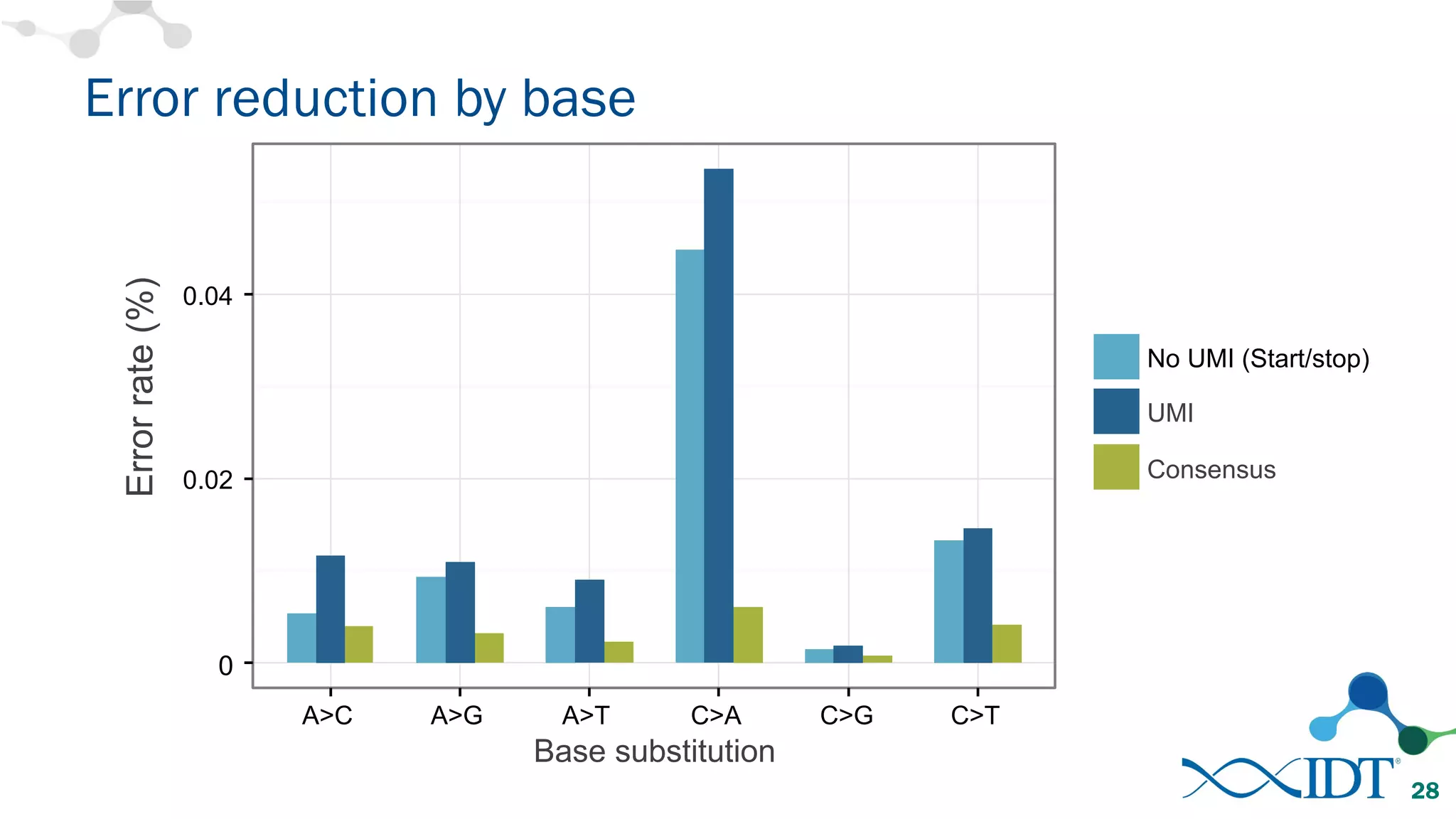
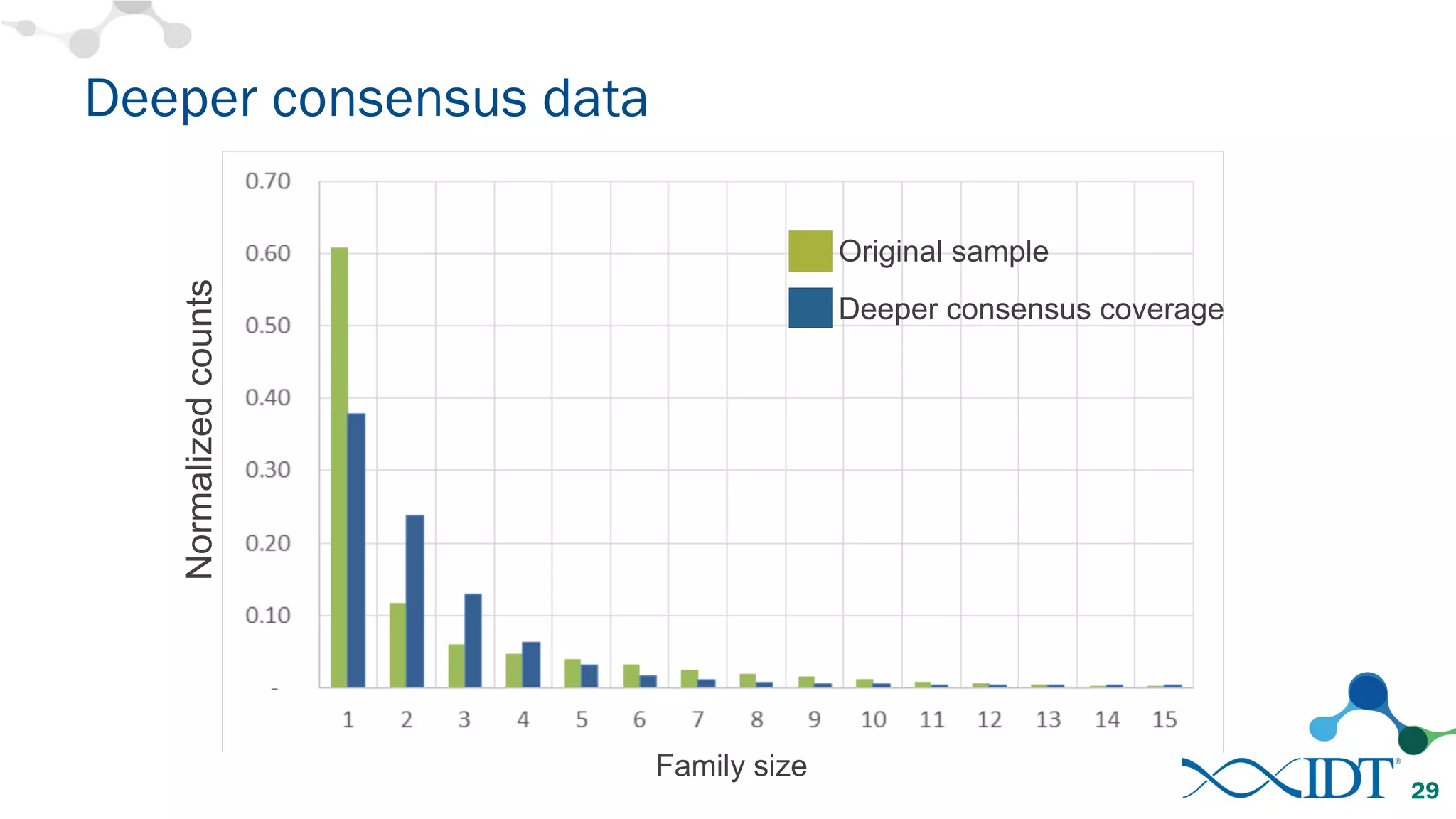
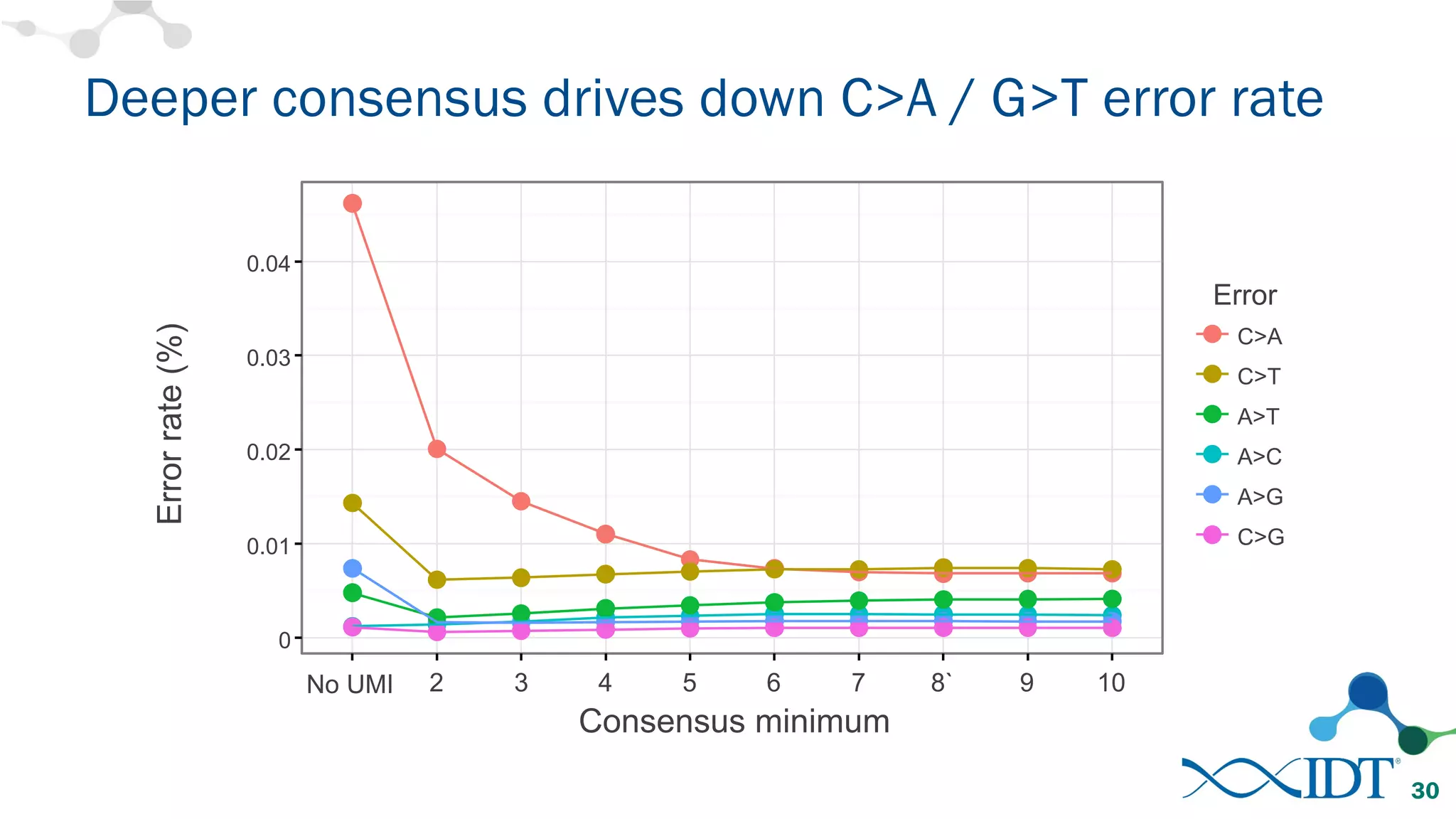
The document discusses the importance of accurately detecting low frequency genetic variants through new molecular tagged sequencing adapters and the challenges associated with liquid biopsies. It outlines experimental results showing how unique molecular identifiers (UMIs) enhance the accuracy of variant detection, leading to significantly lower false positives. The conclusions emphasize that utilizing UMIs in sequencing libraries improves overall sensitivity and positive predictive value for variant calling down to 0.25% frequency.