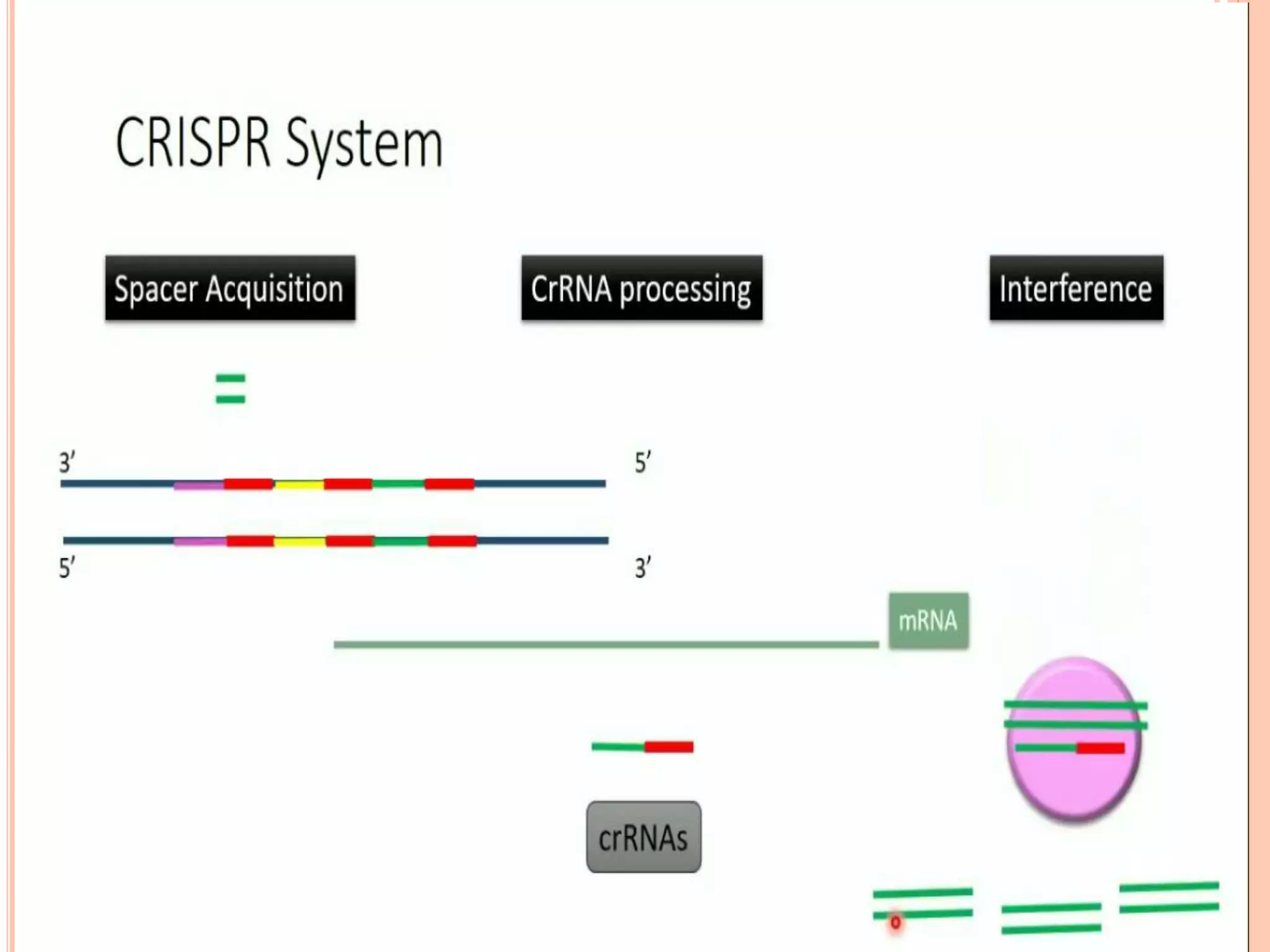
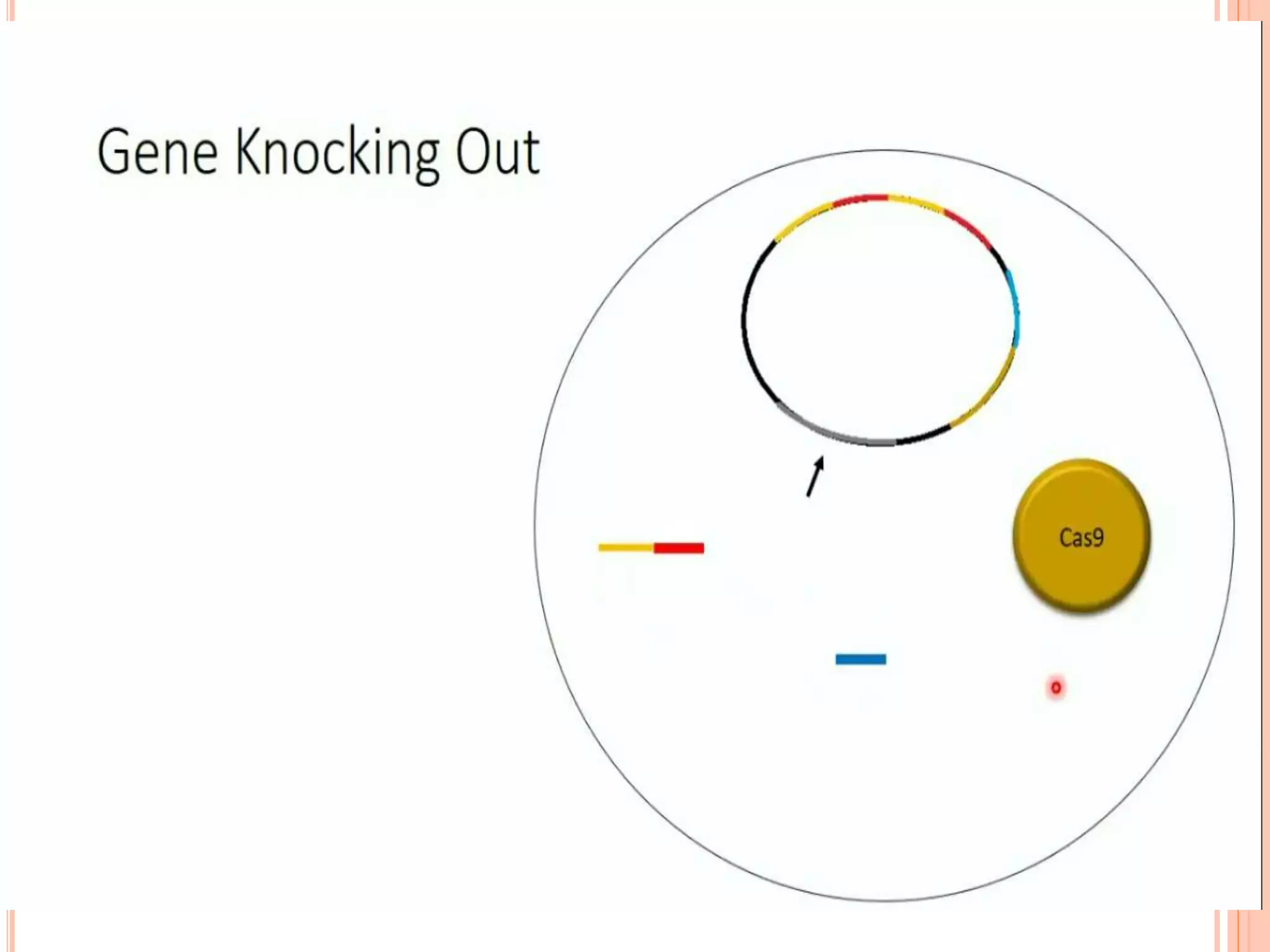
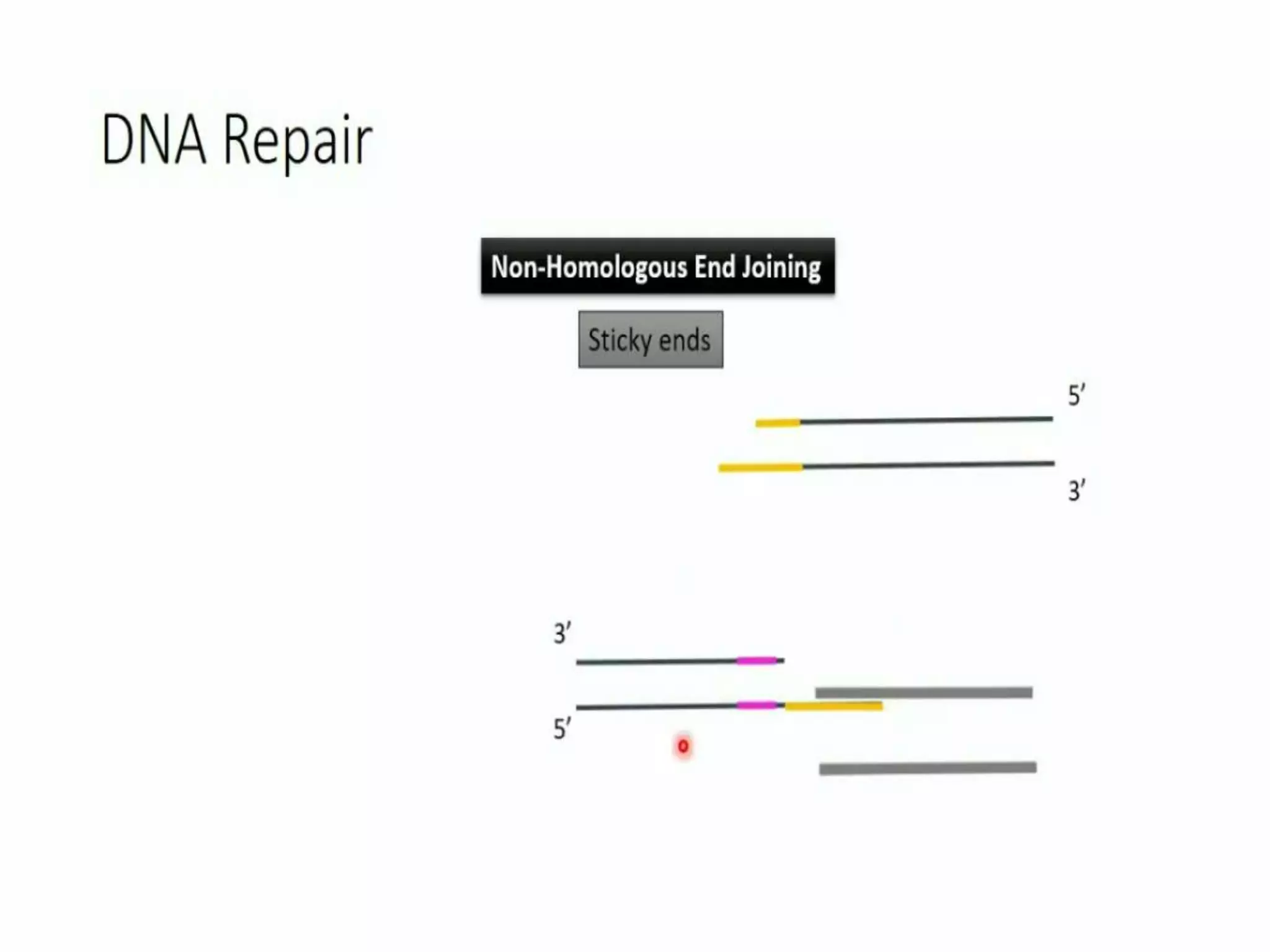
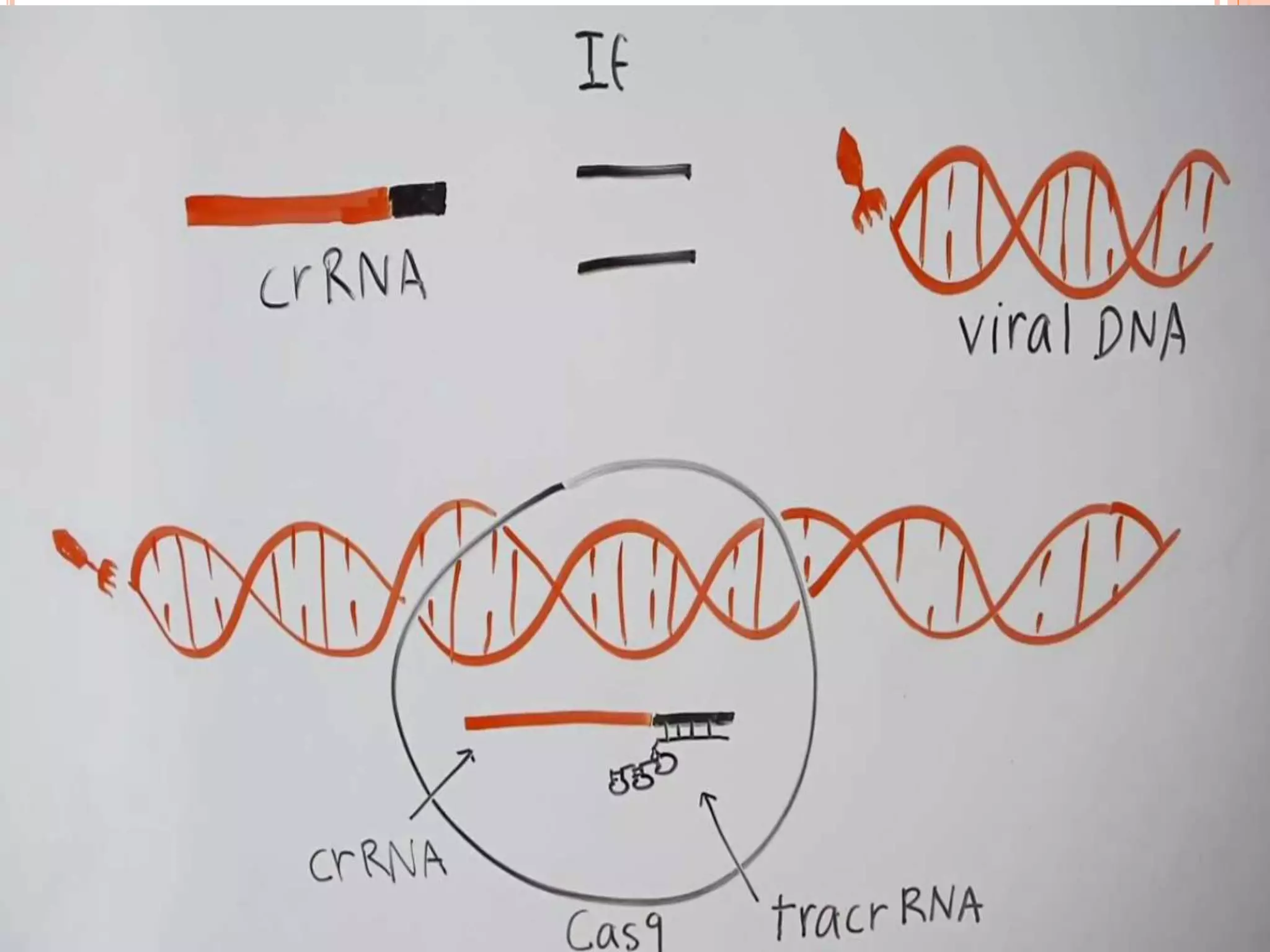
This document discusses genome editing using the CRISPR-Cas9 system. It begins by introducing three main genome editing technologies - zinc-finger nucleases, TALENs, and the CRISPR-Cas9 system. It then describes the key events in the discovery of CRISPR-Cas9, including its origins as a bacterial defense system. The document outlines the main components of the CRISPR-Cas9 system, including crRNA, tracrRNA, sgRNA, and Cas9. It also summarizes the two main steps in genome editing using CRISPR-Cas9 - knocking out genes and DNA repair. The document concludes by discussing opportunities for applying CRISPR-Cas9 technology across various