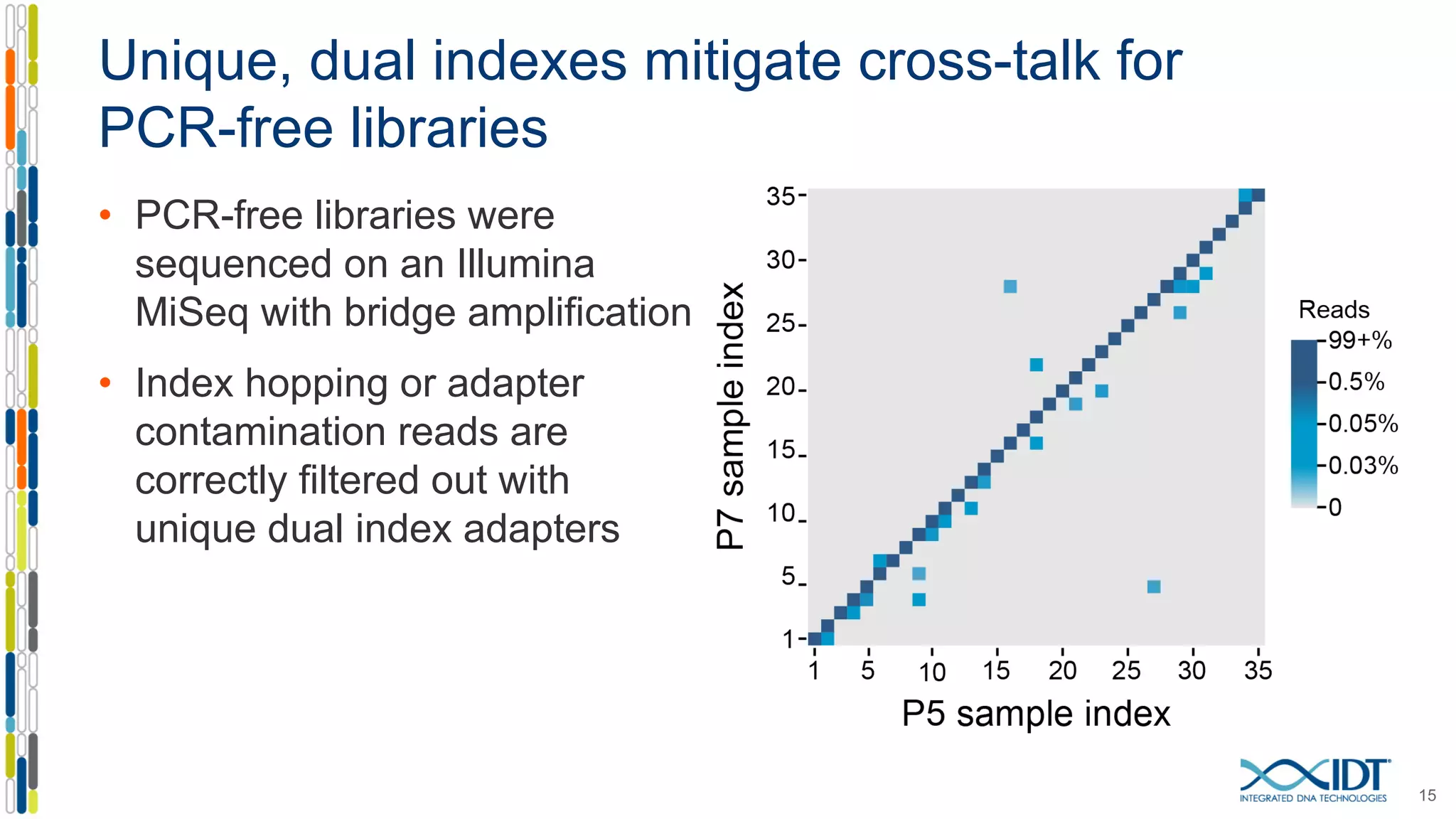
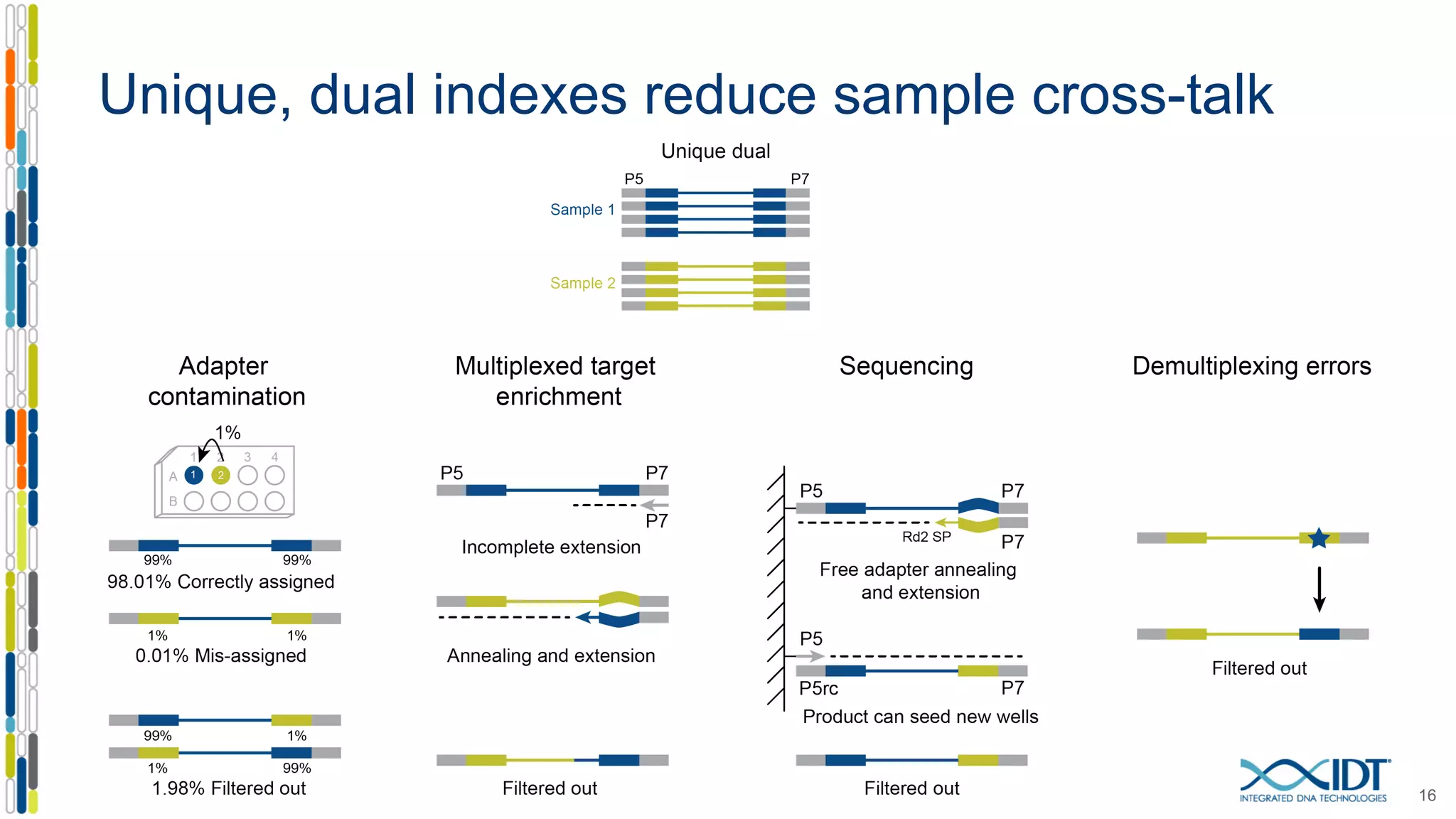
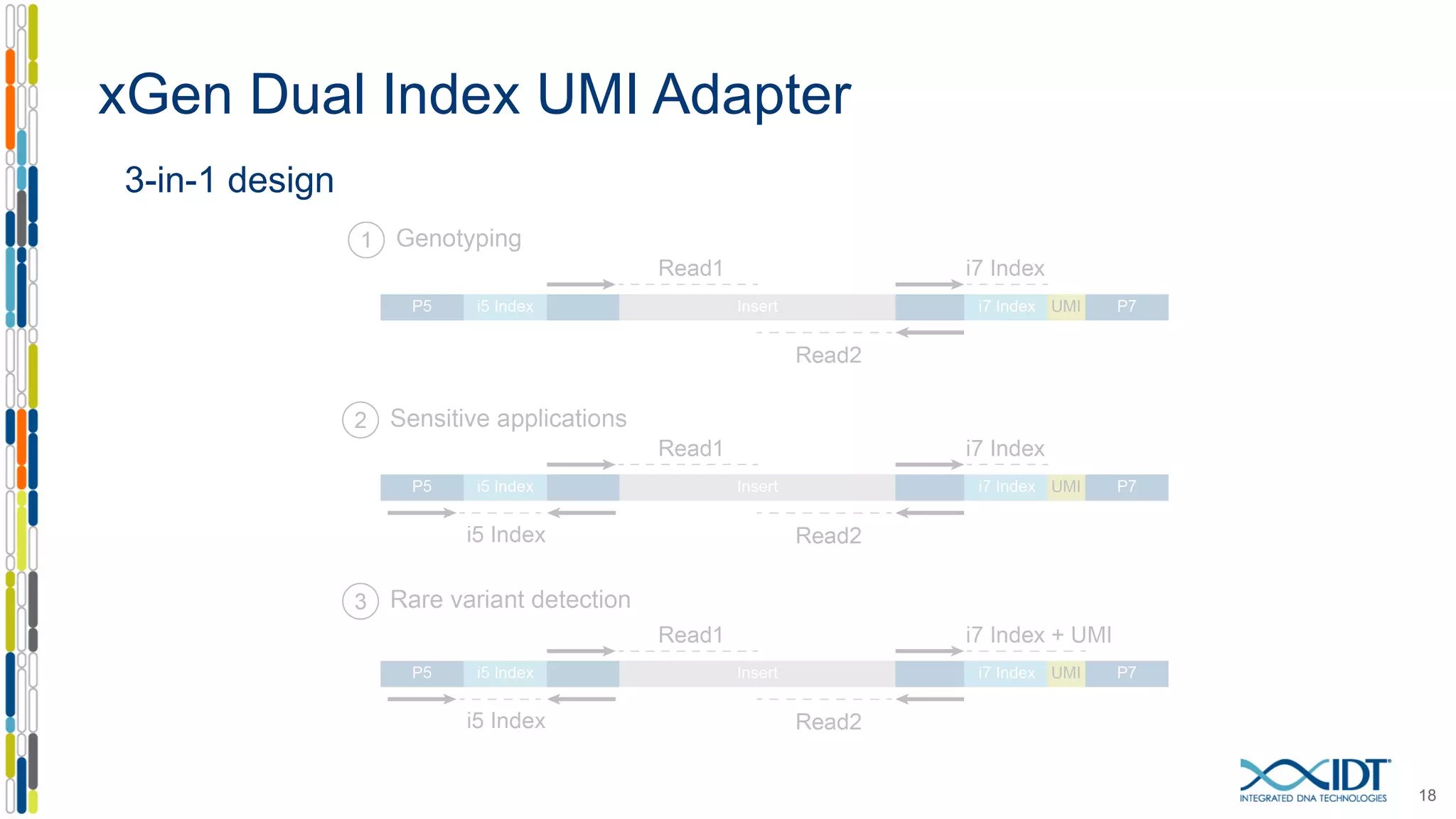
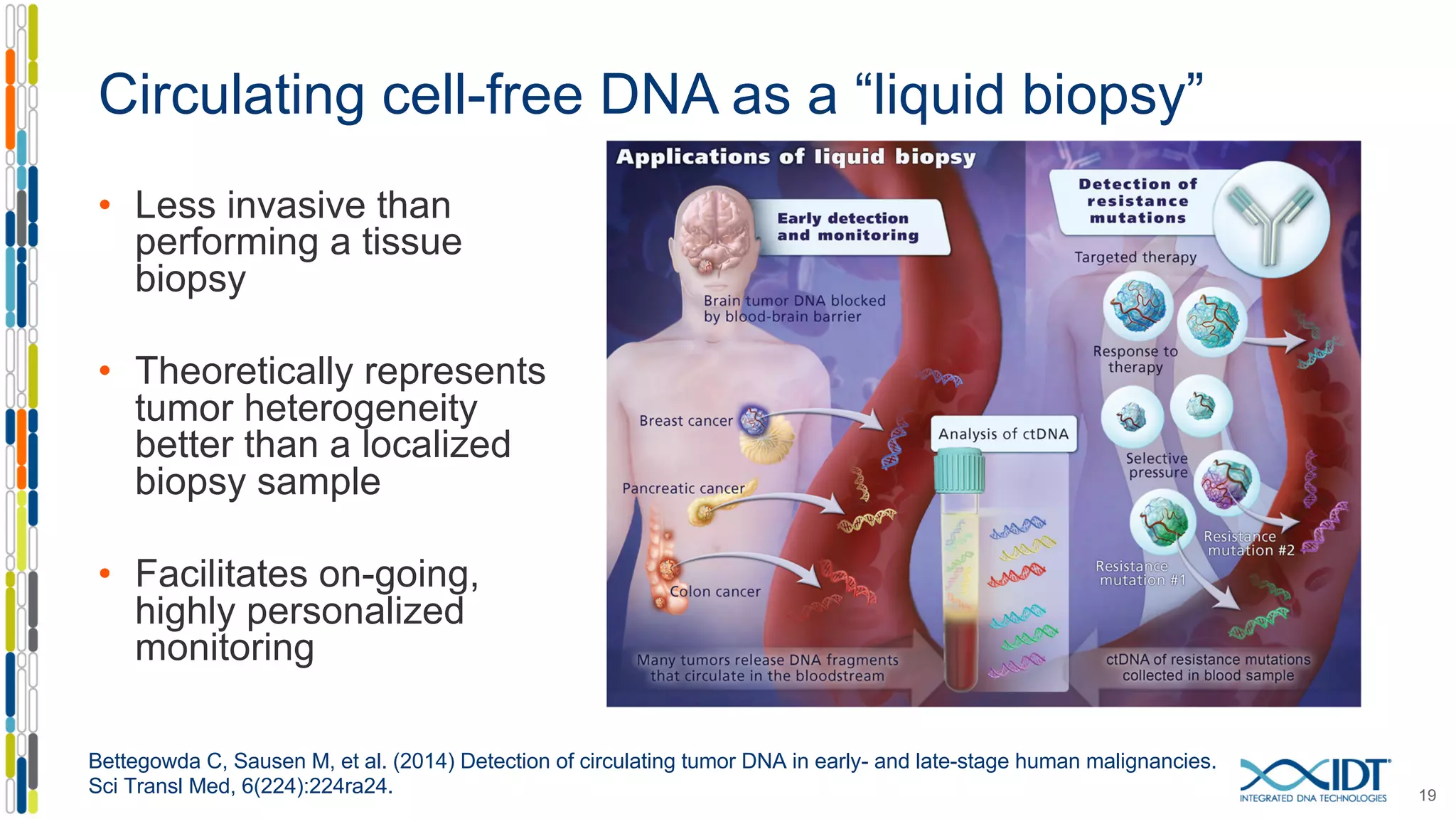
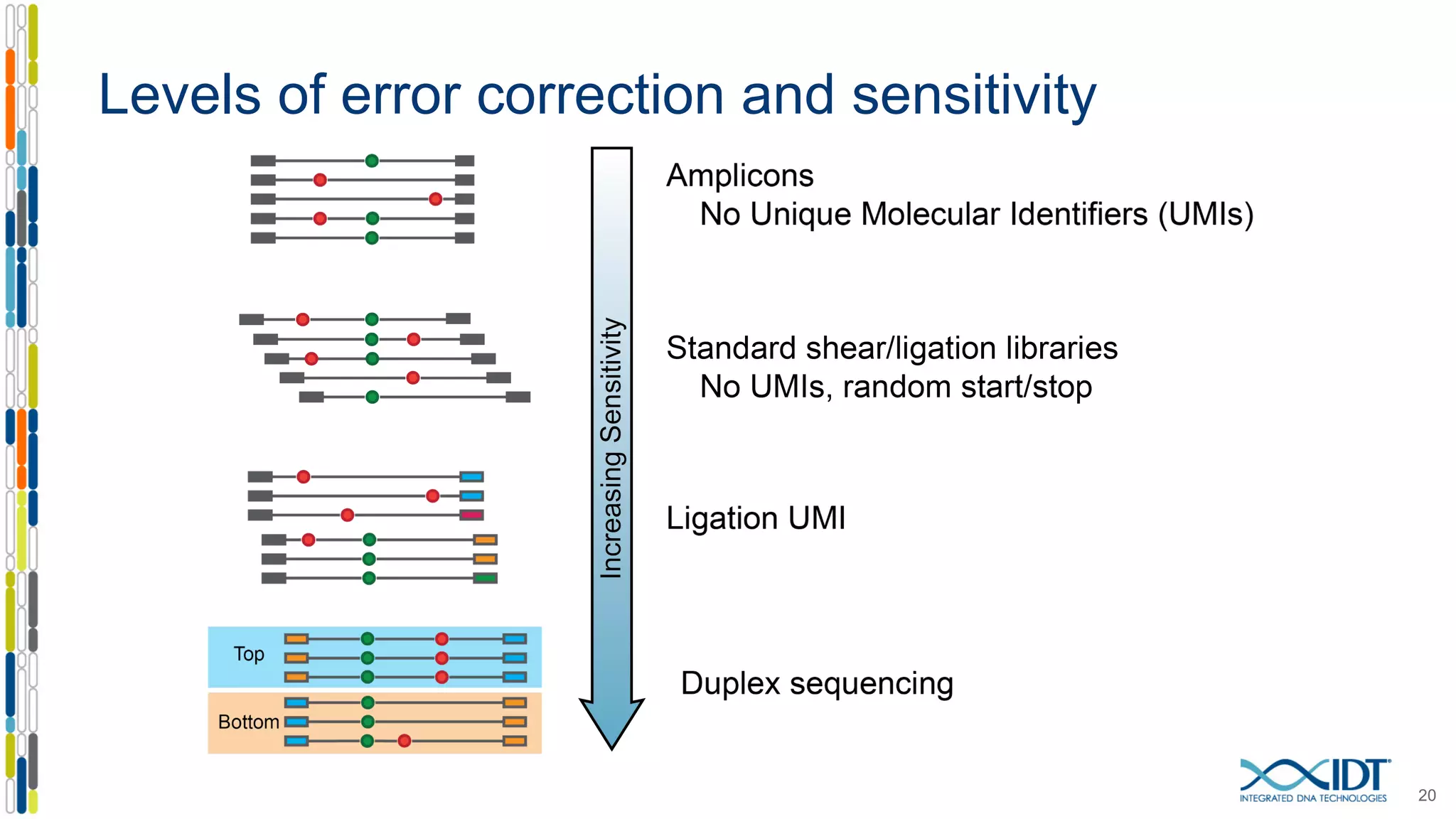
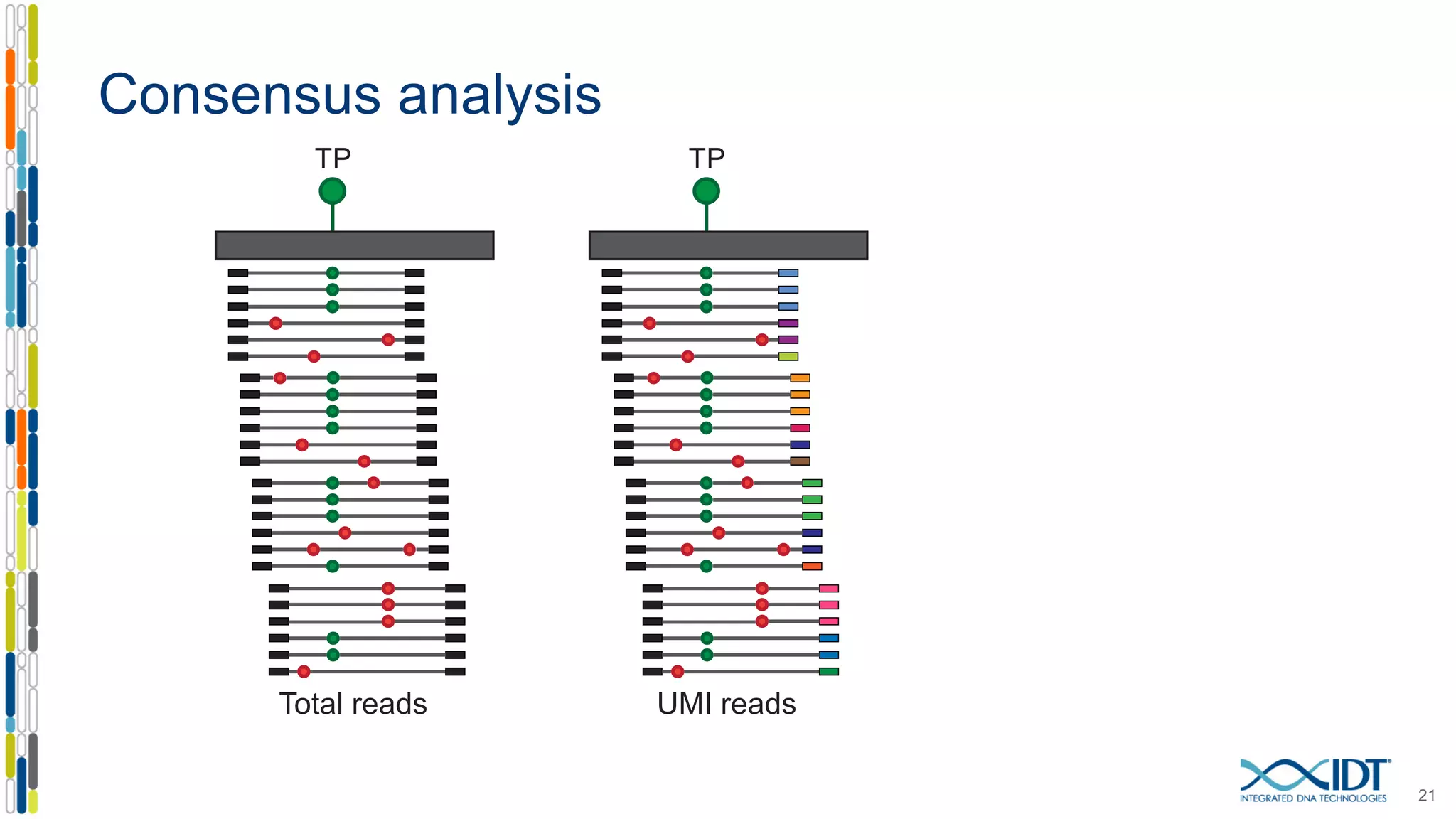
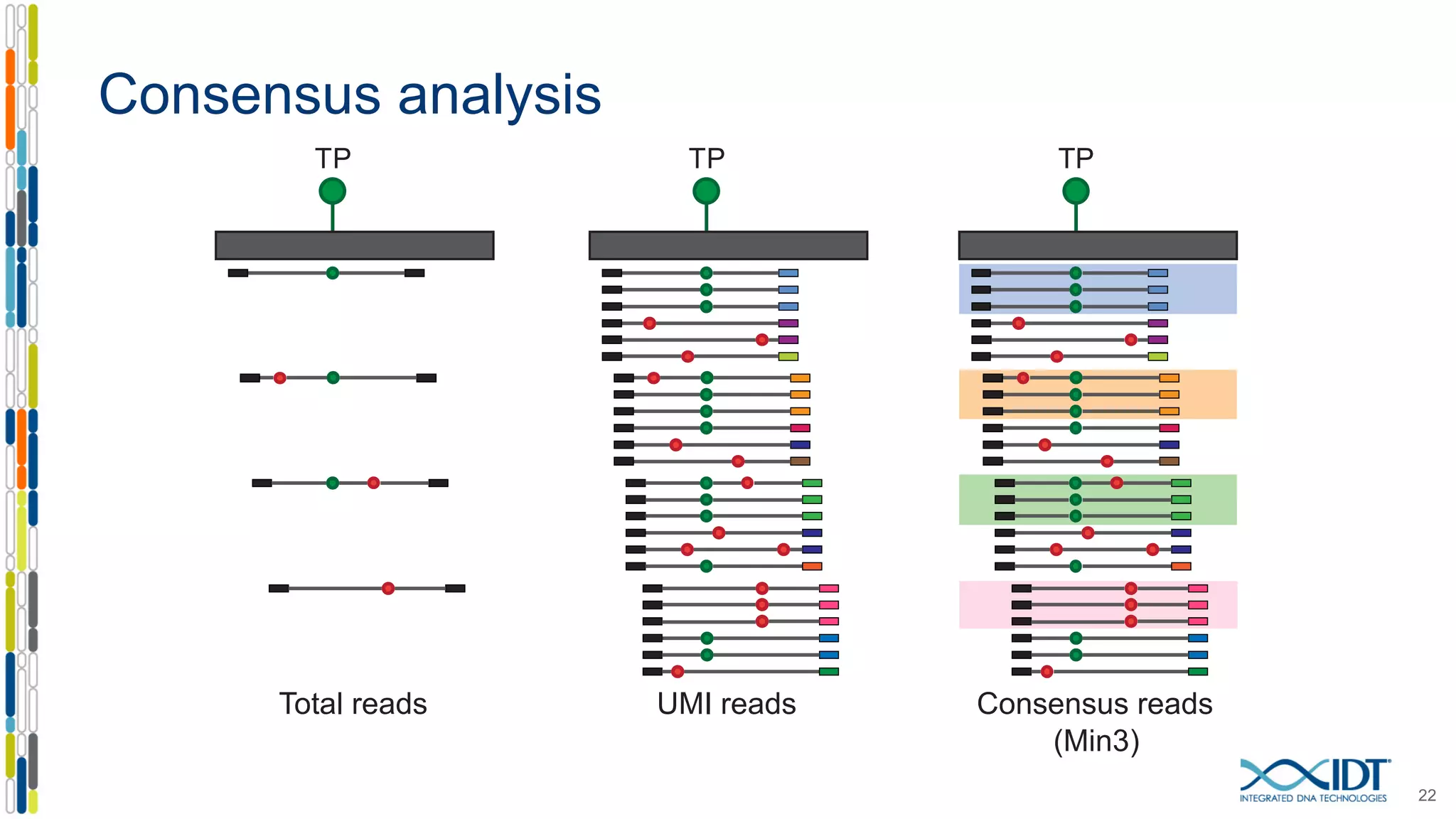
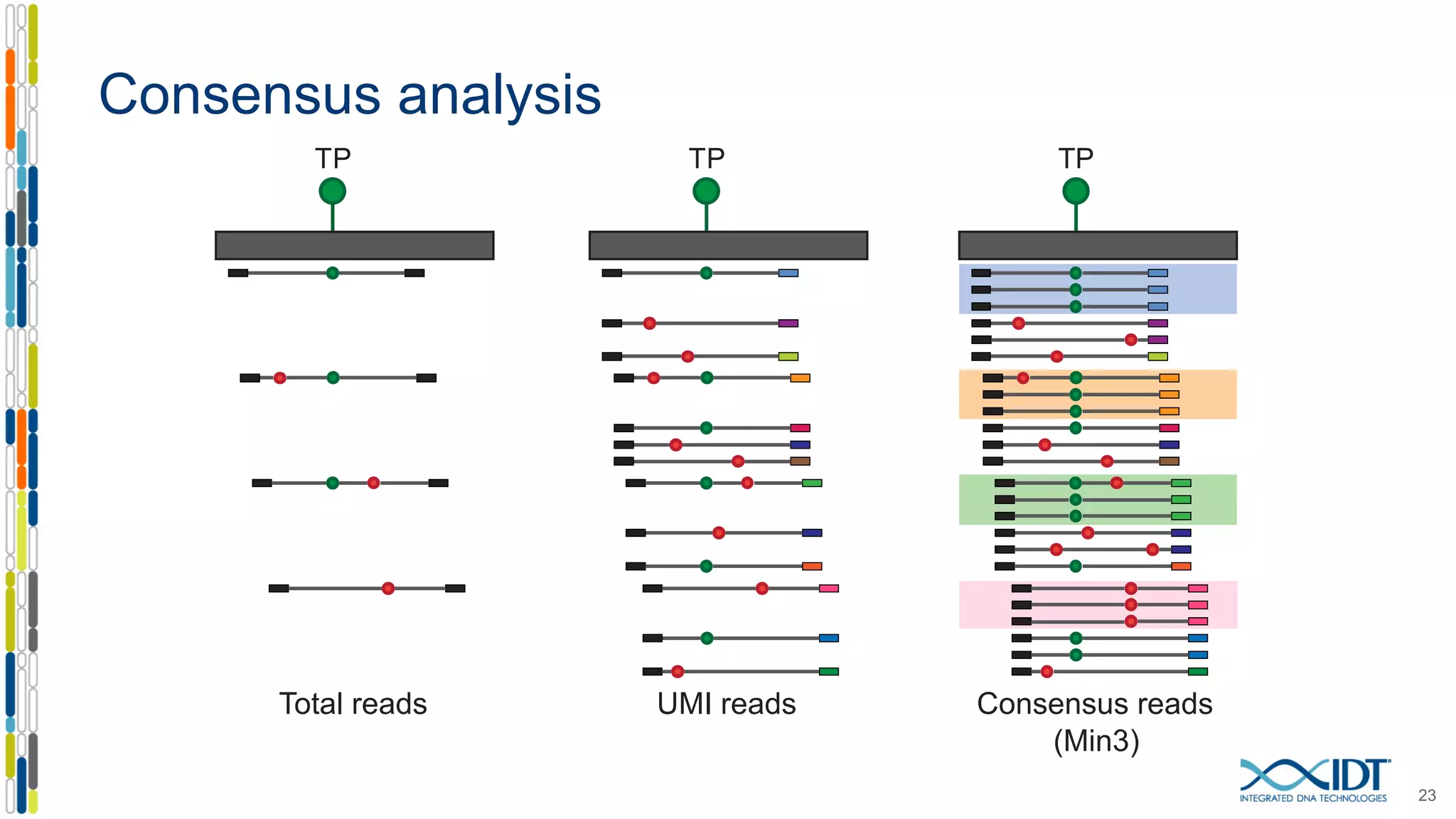
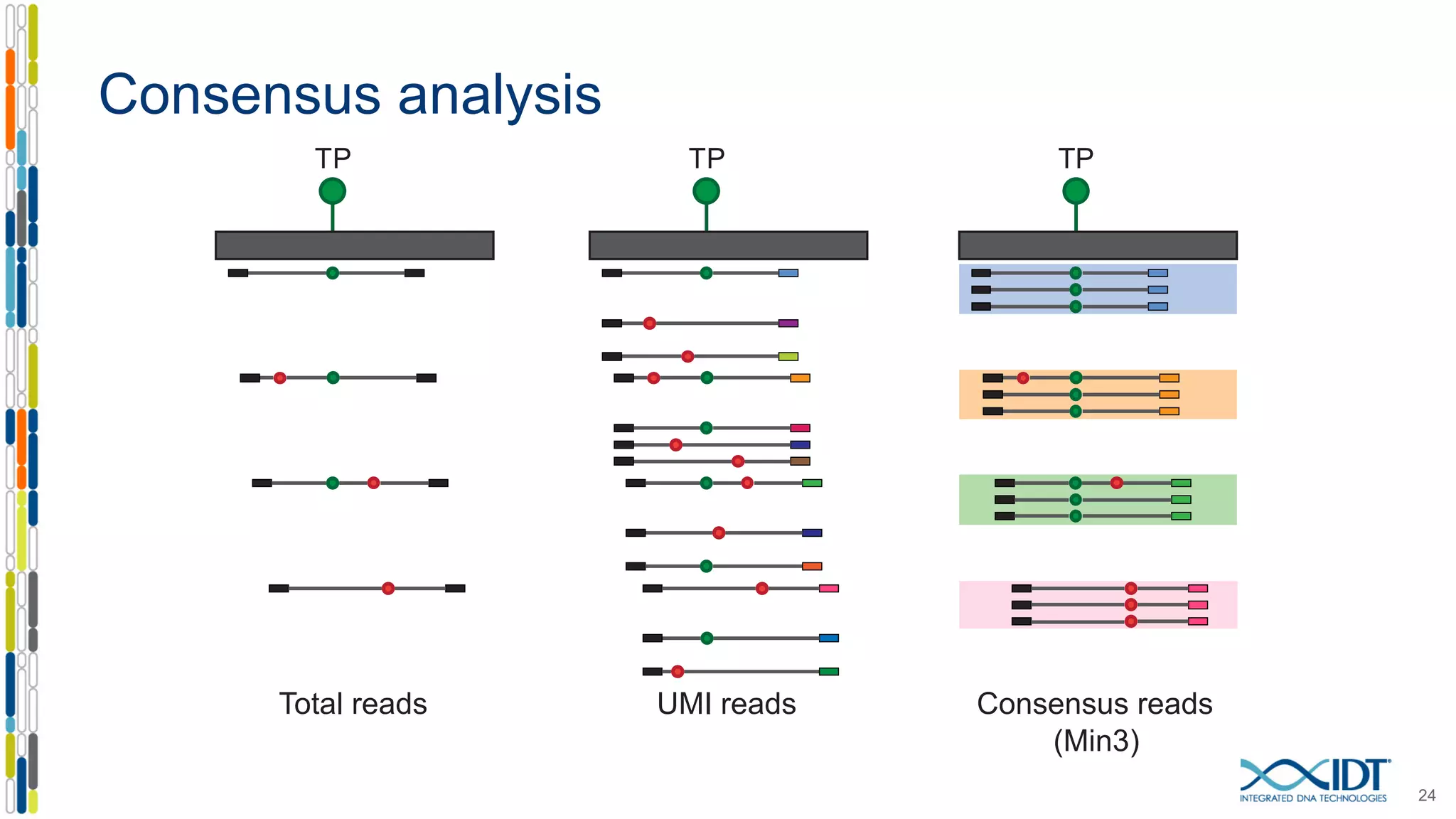
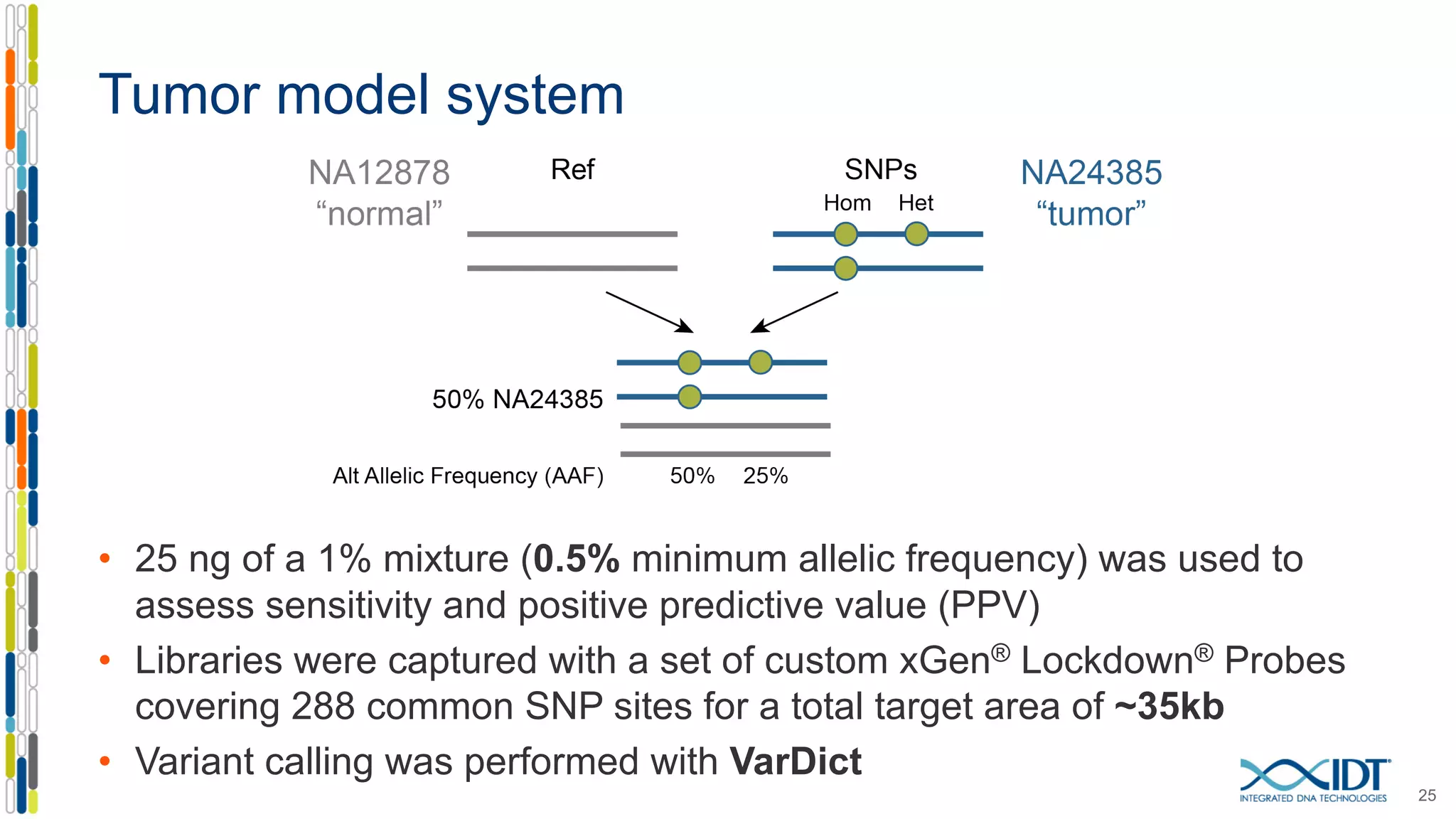
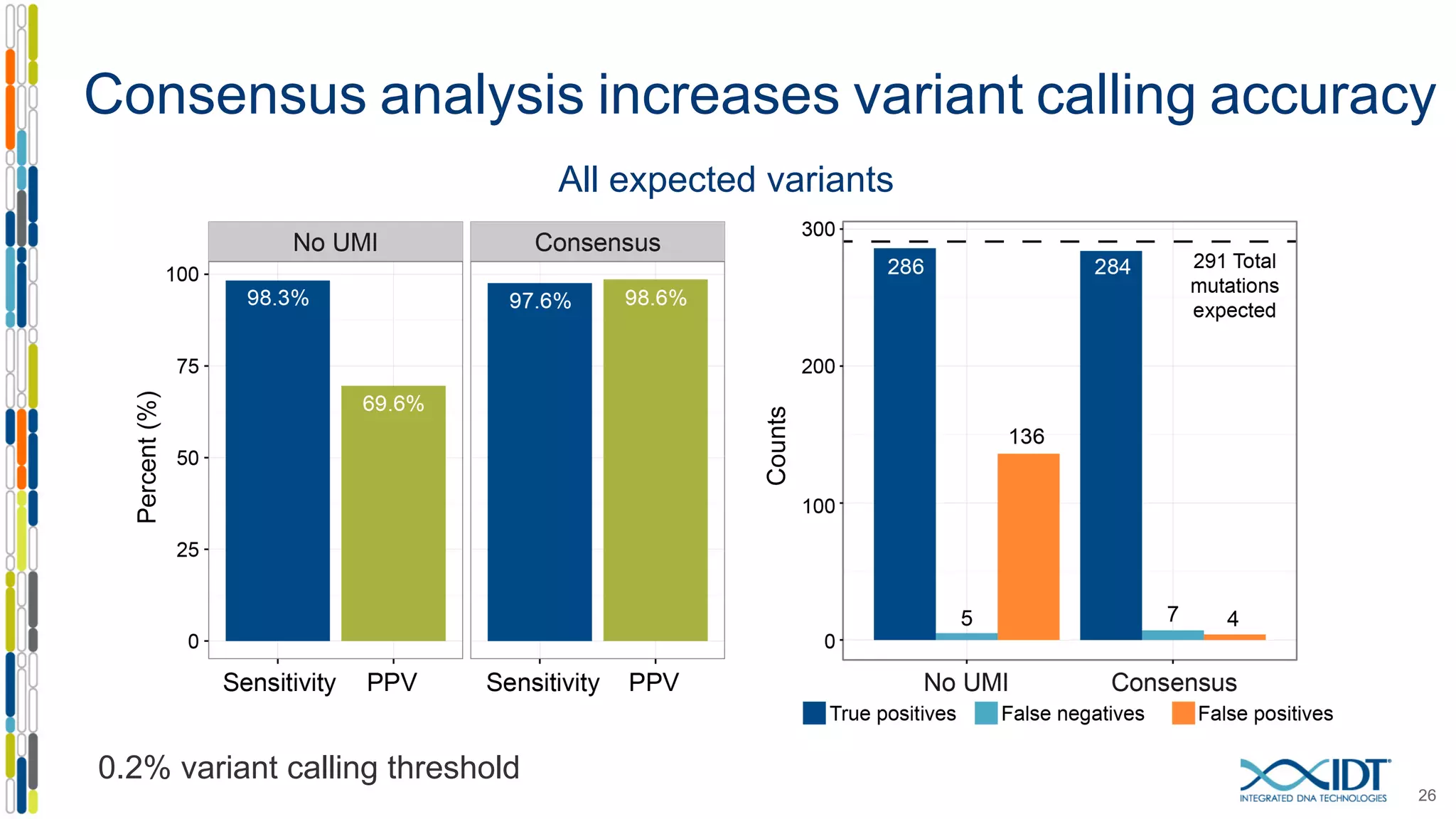
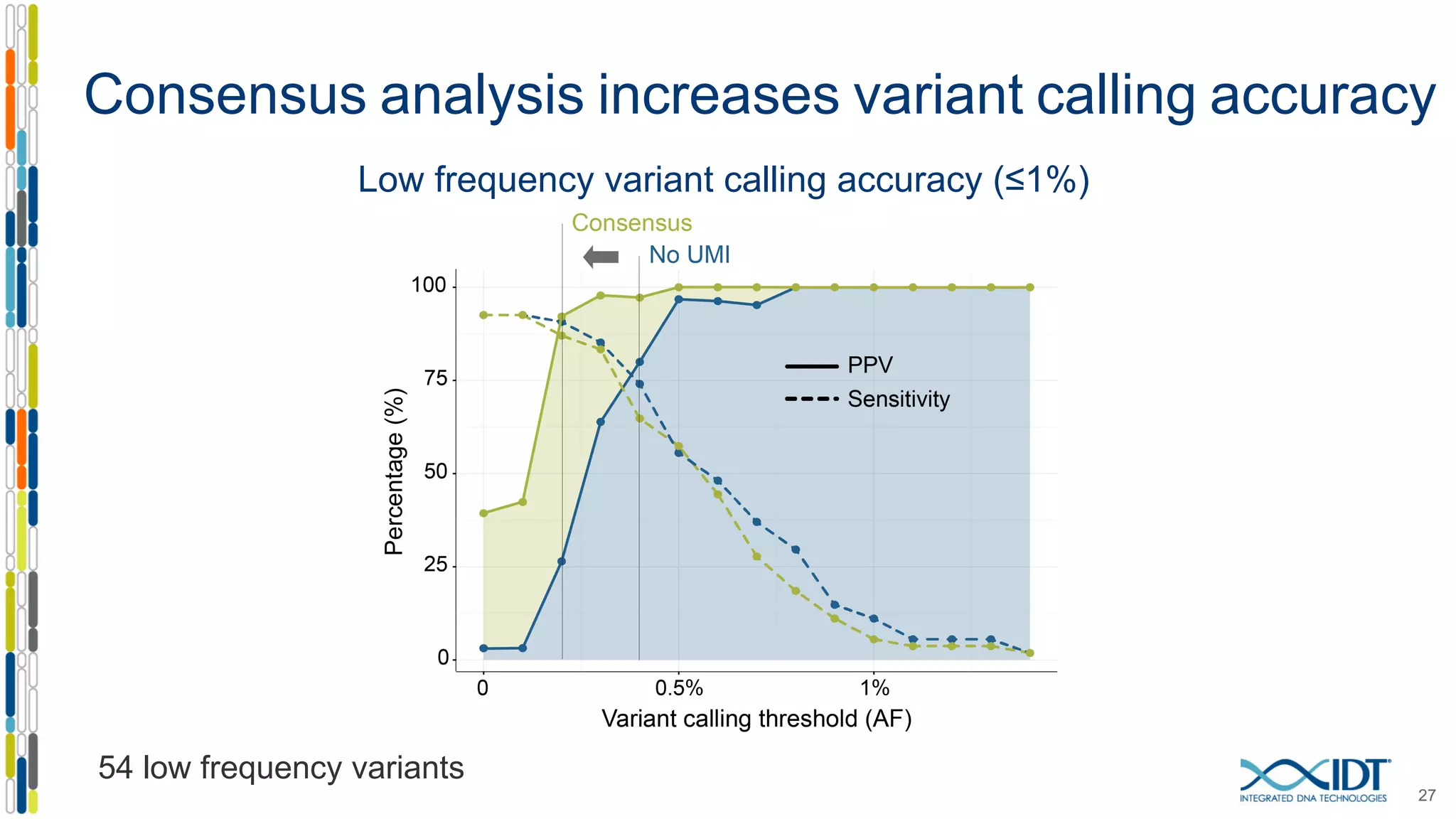
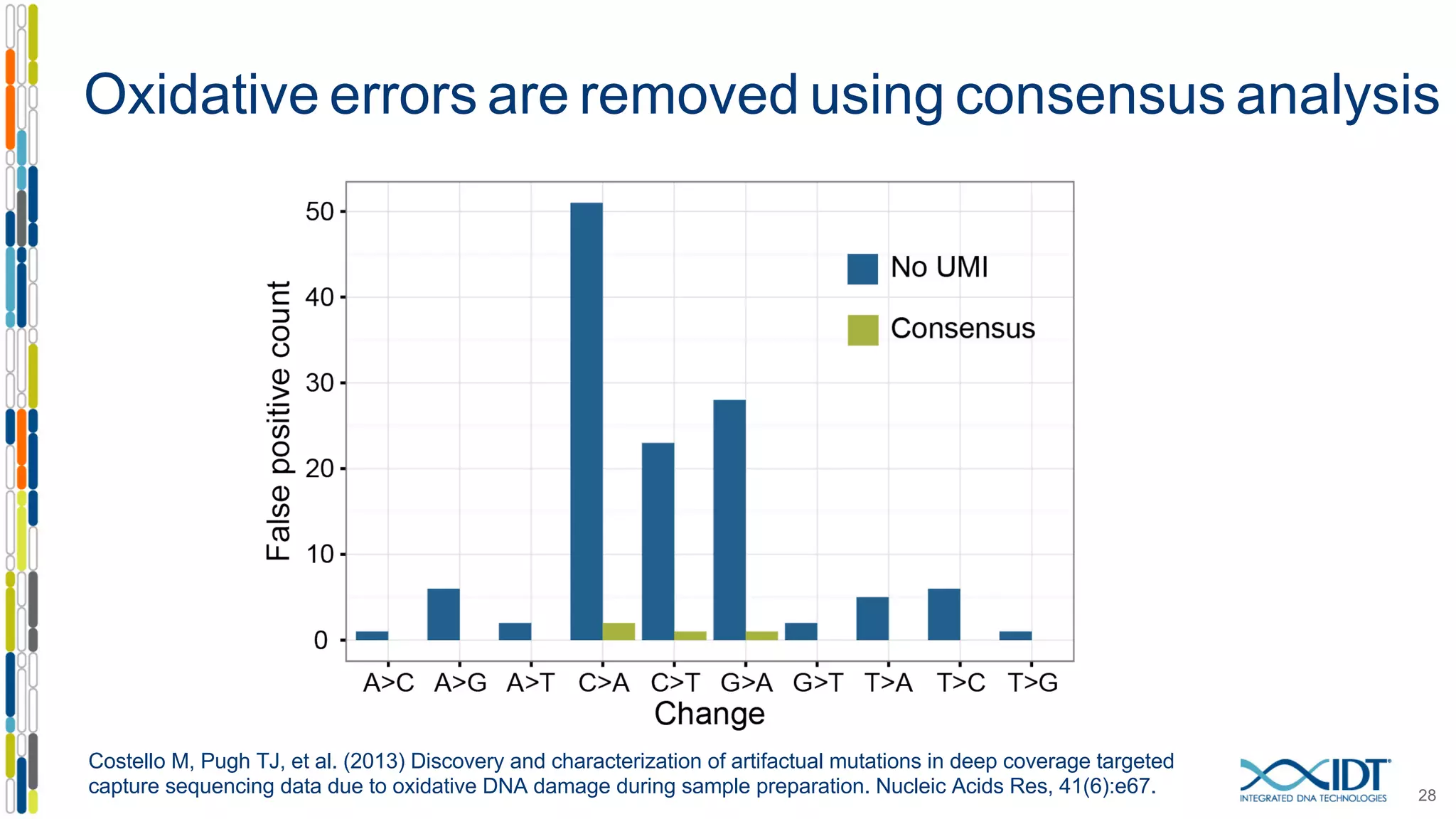
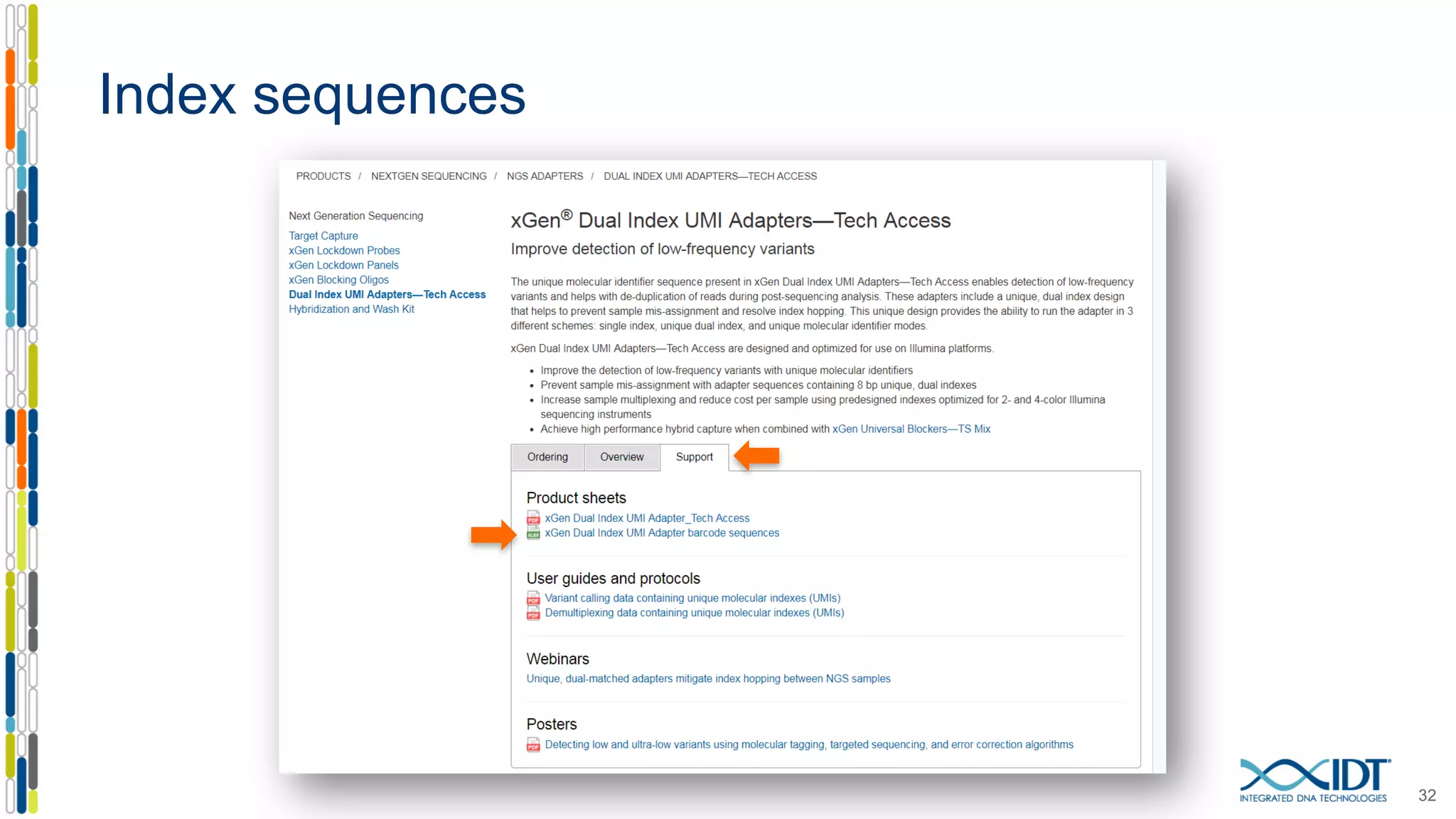
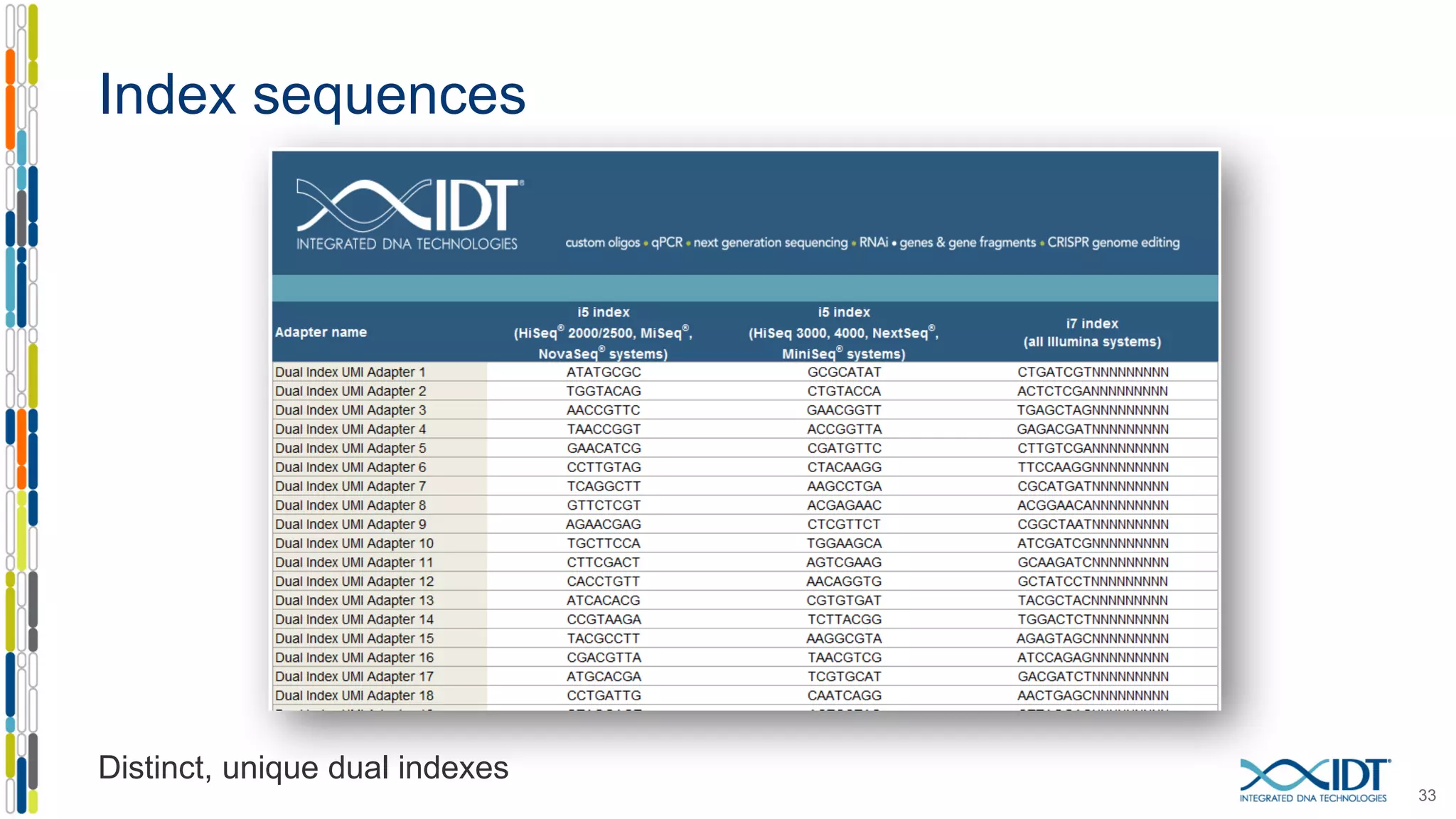
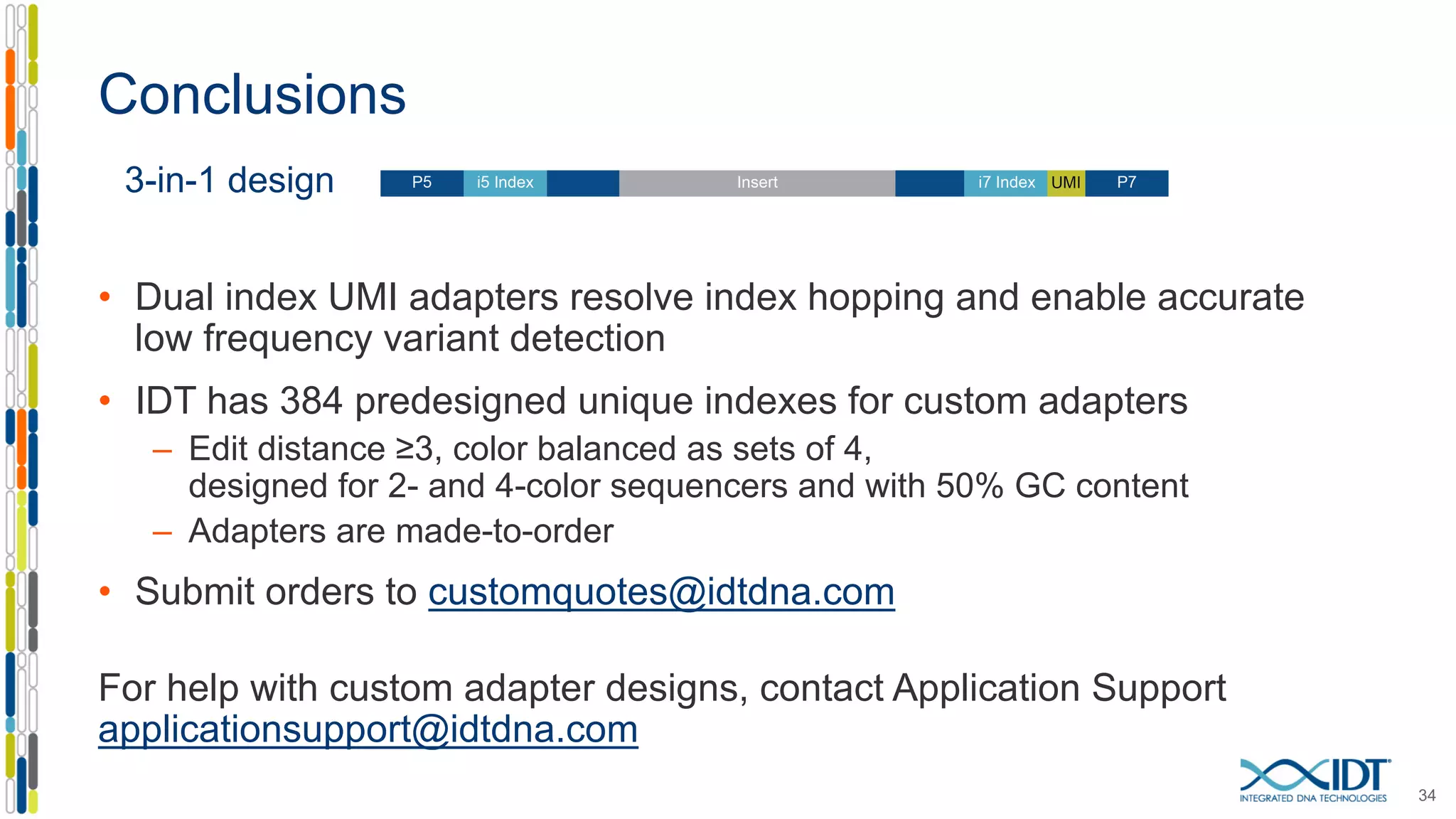
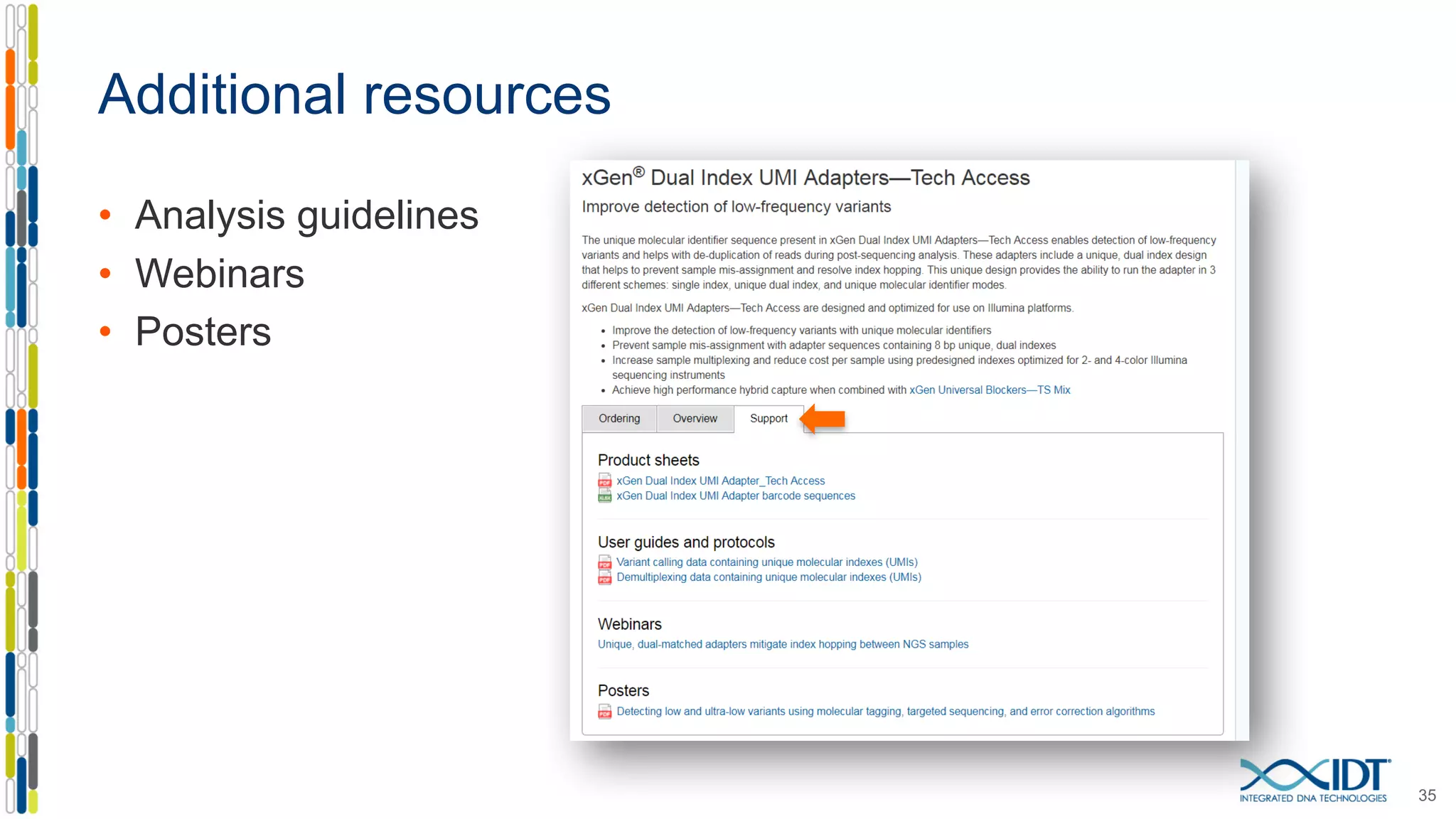
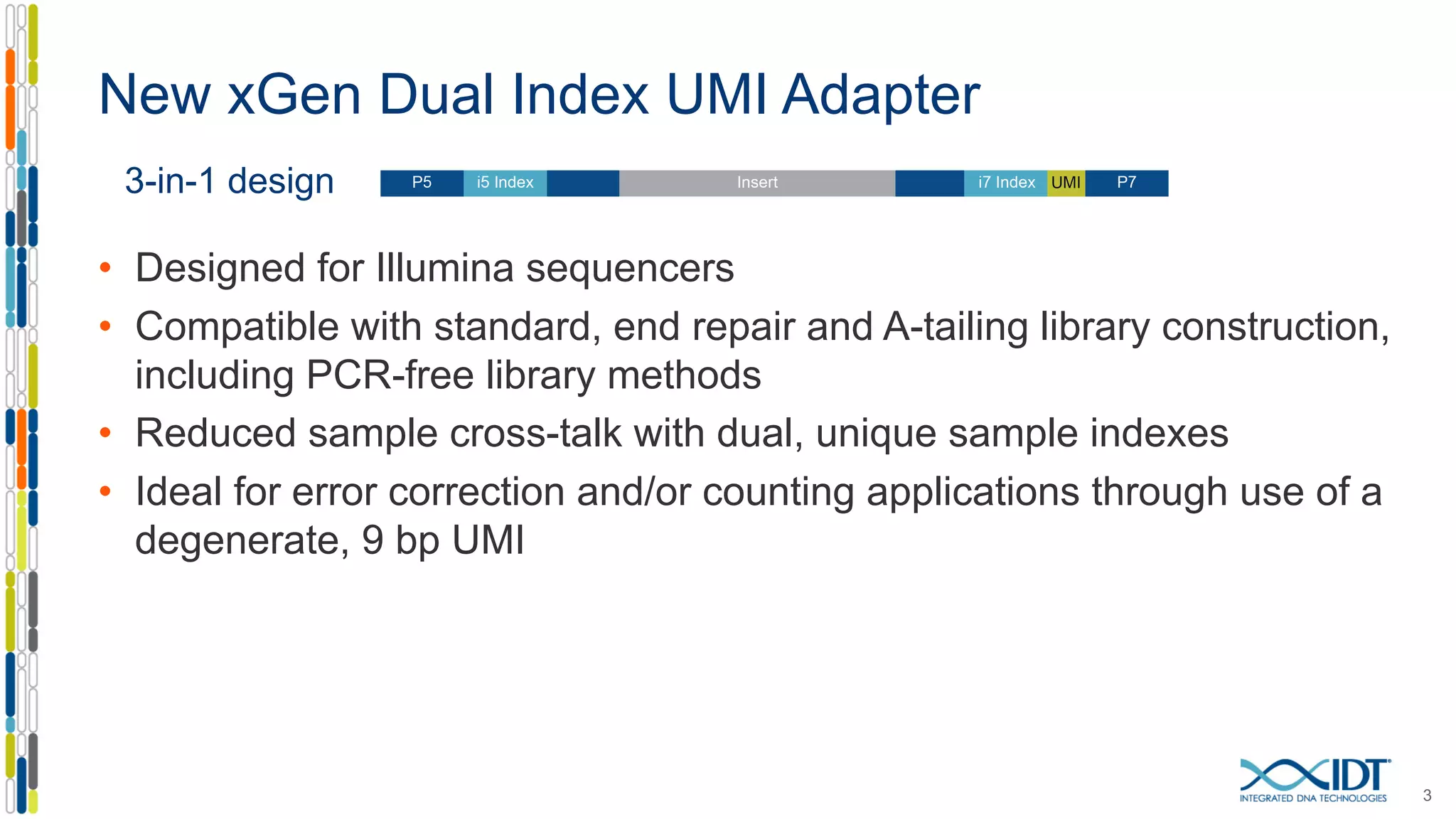
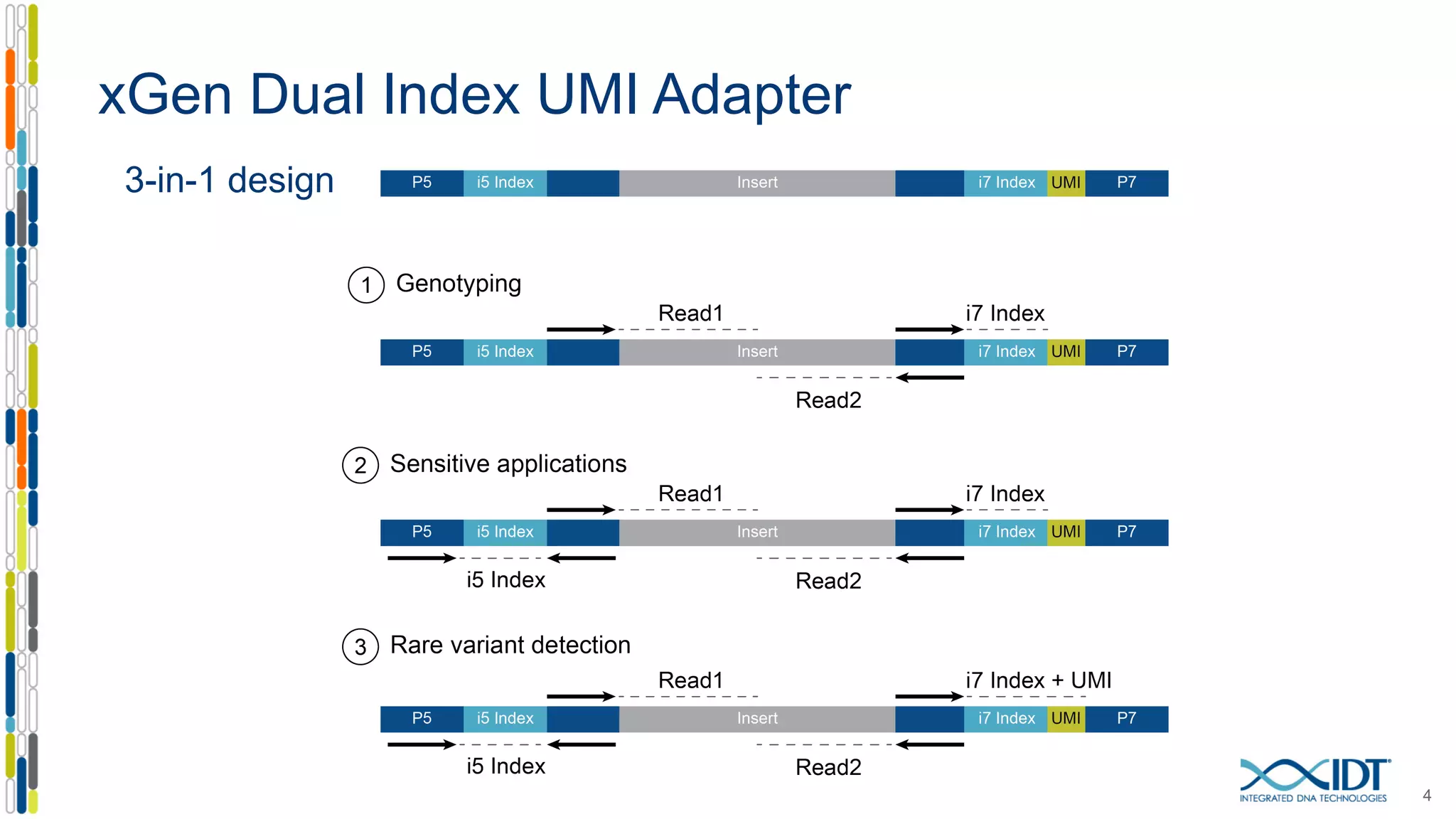
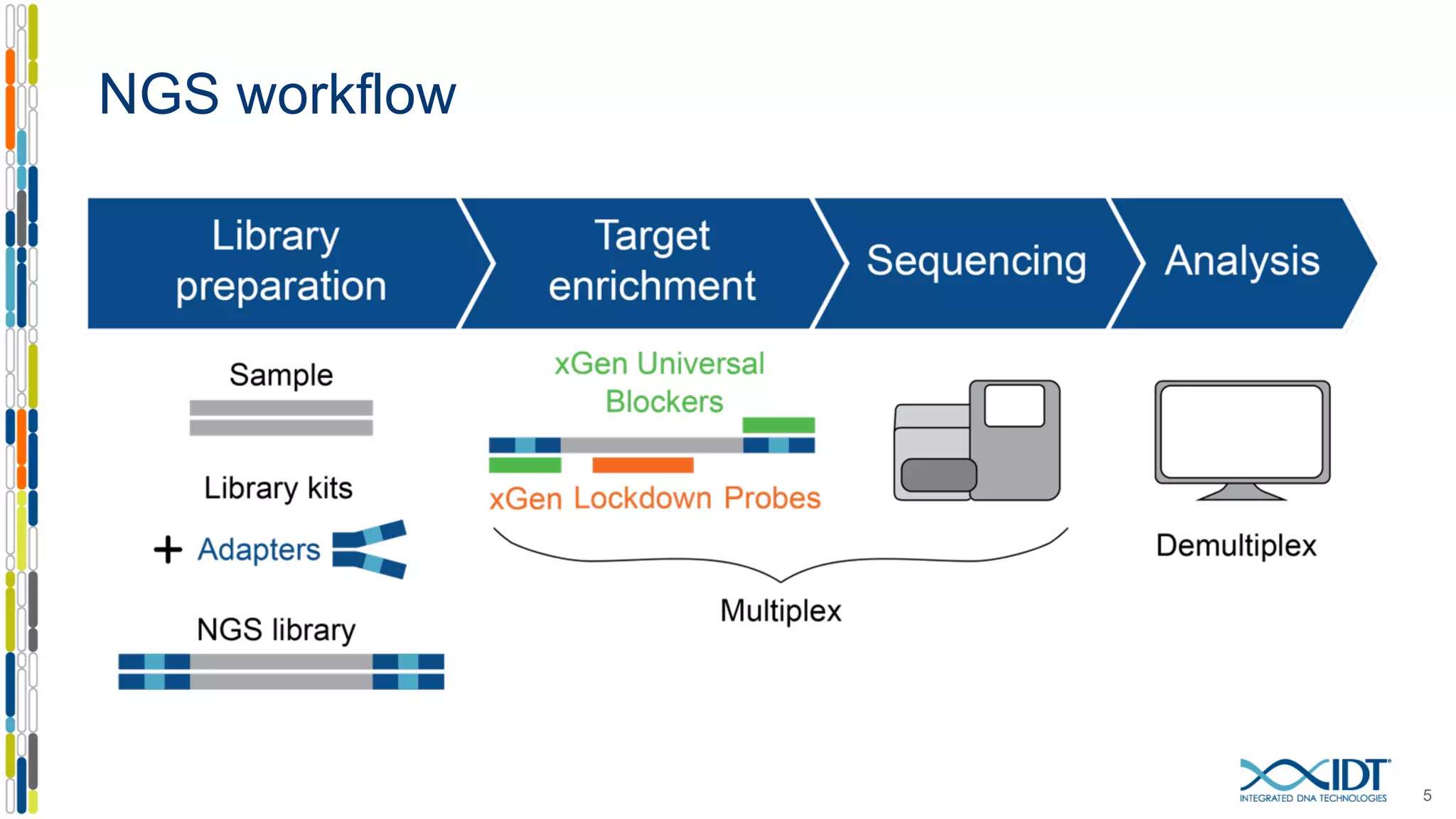
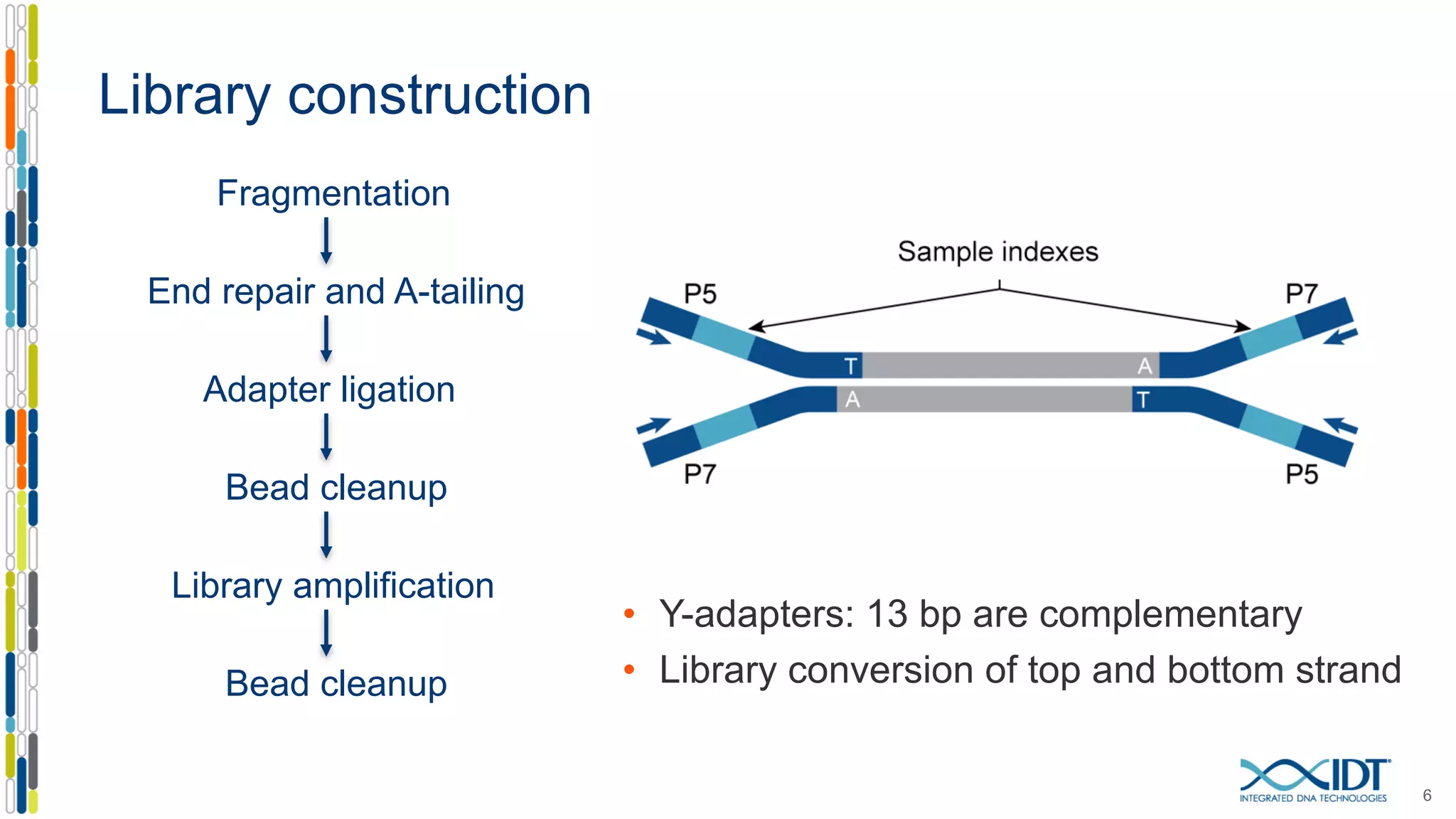
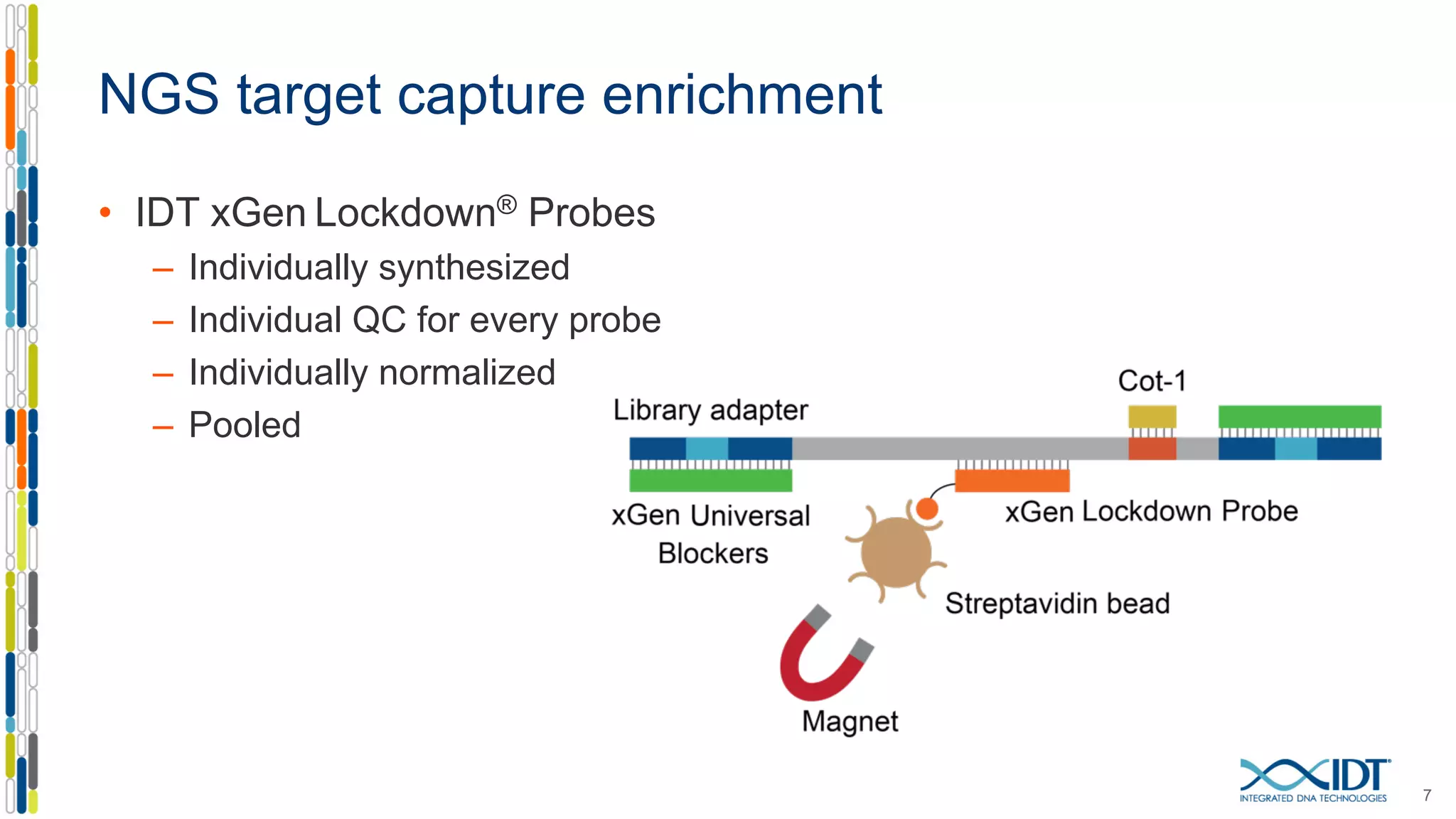
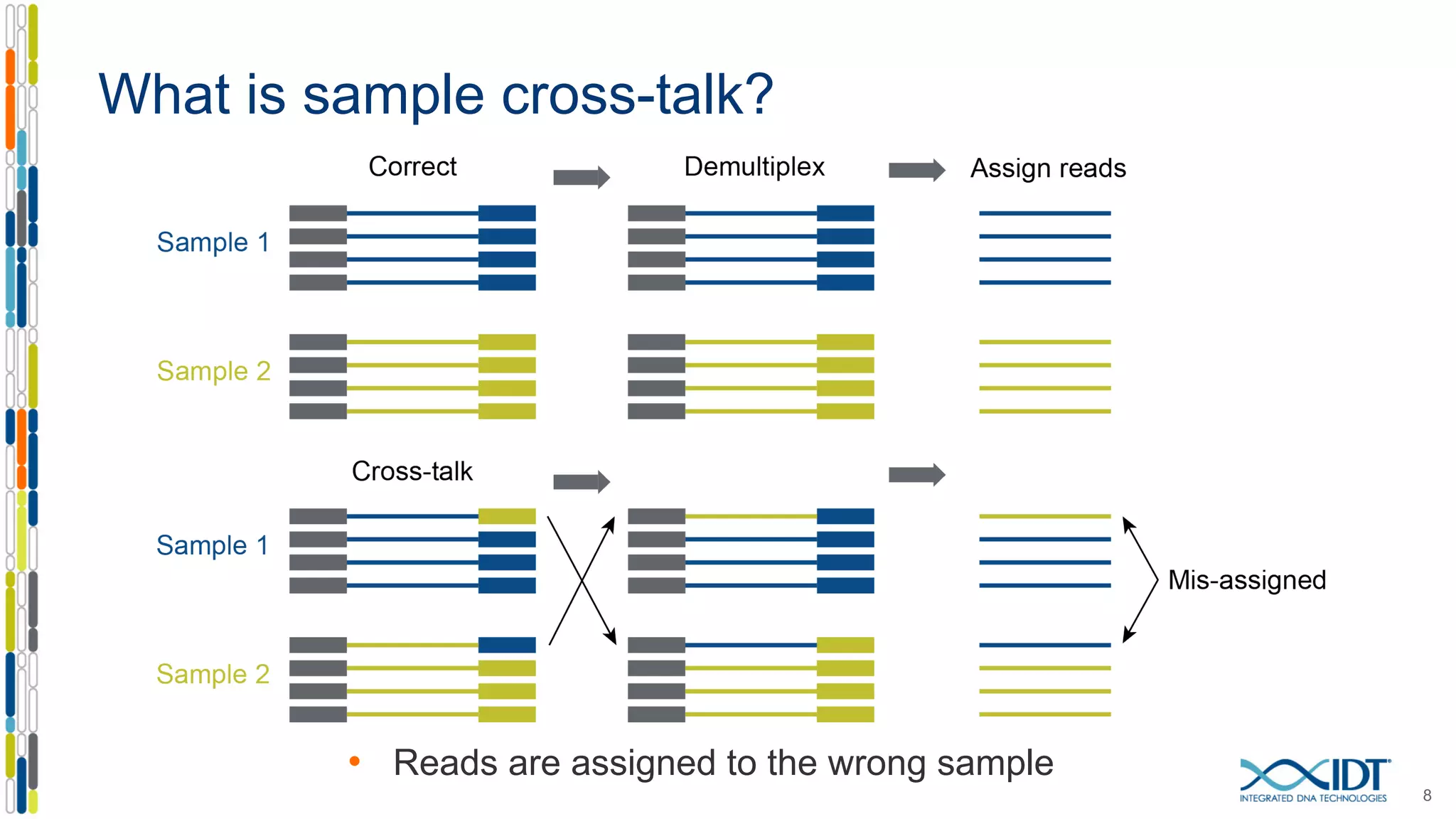
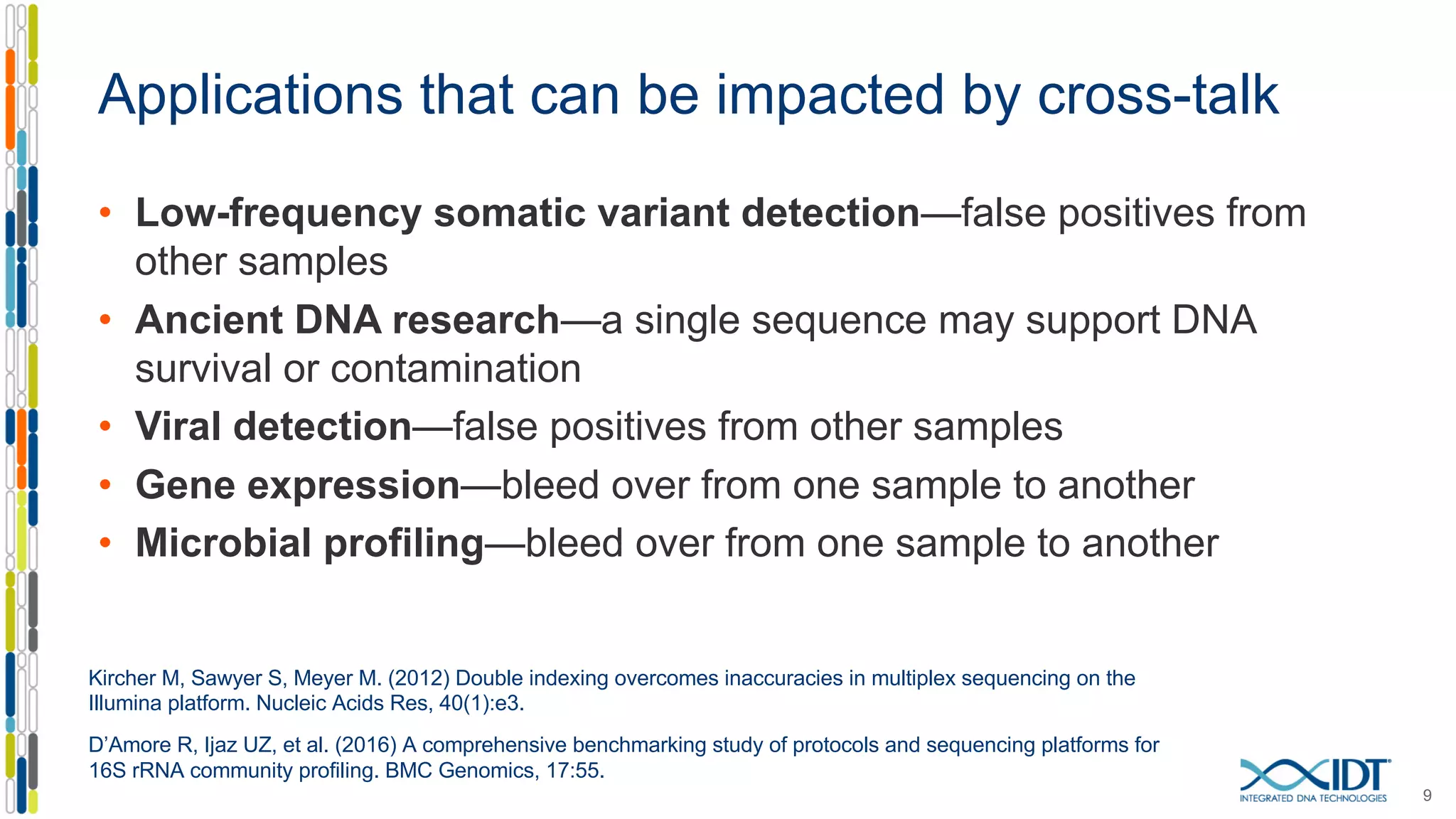
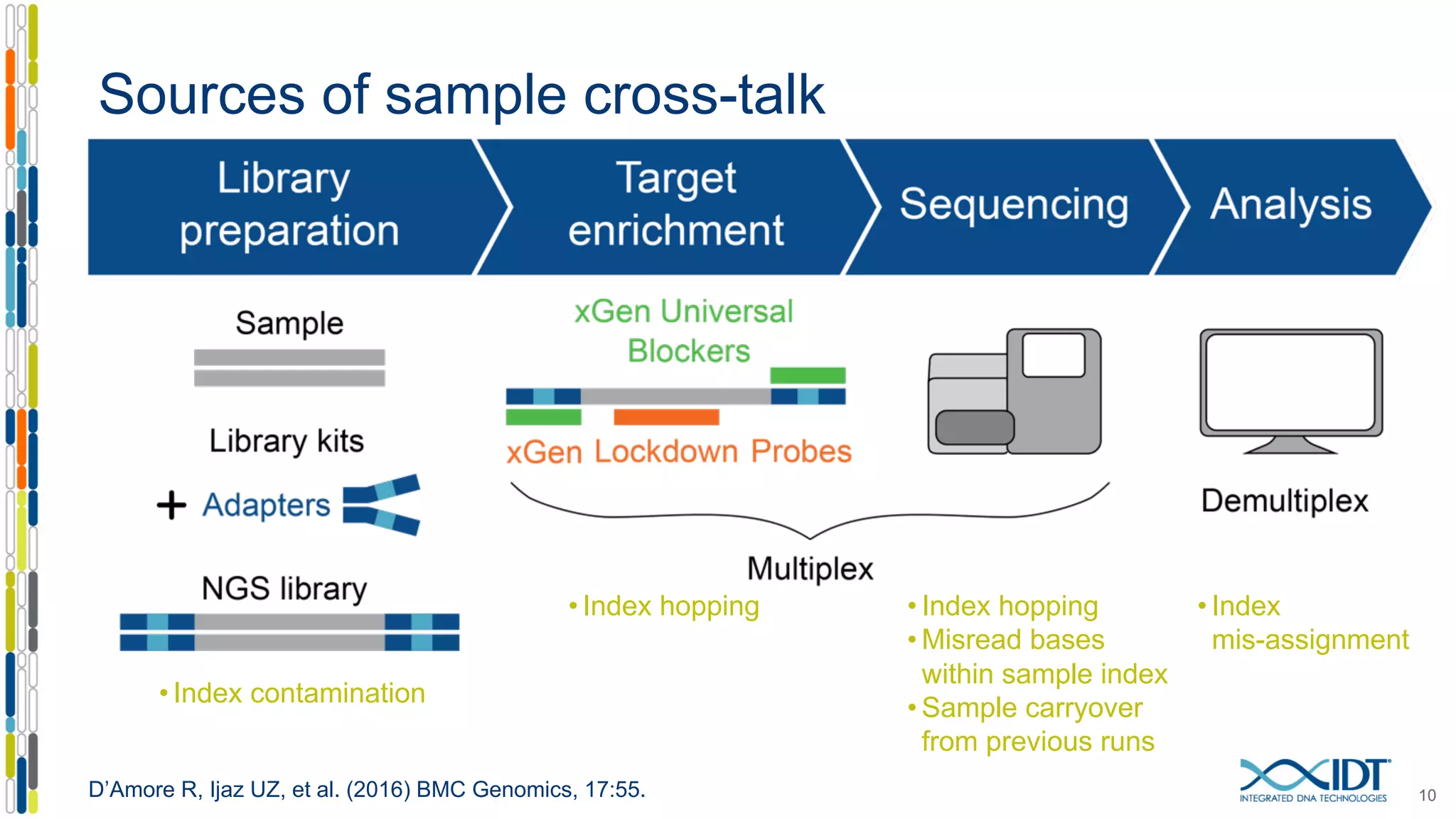
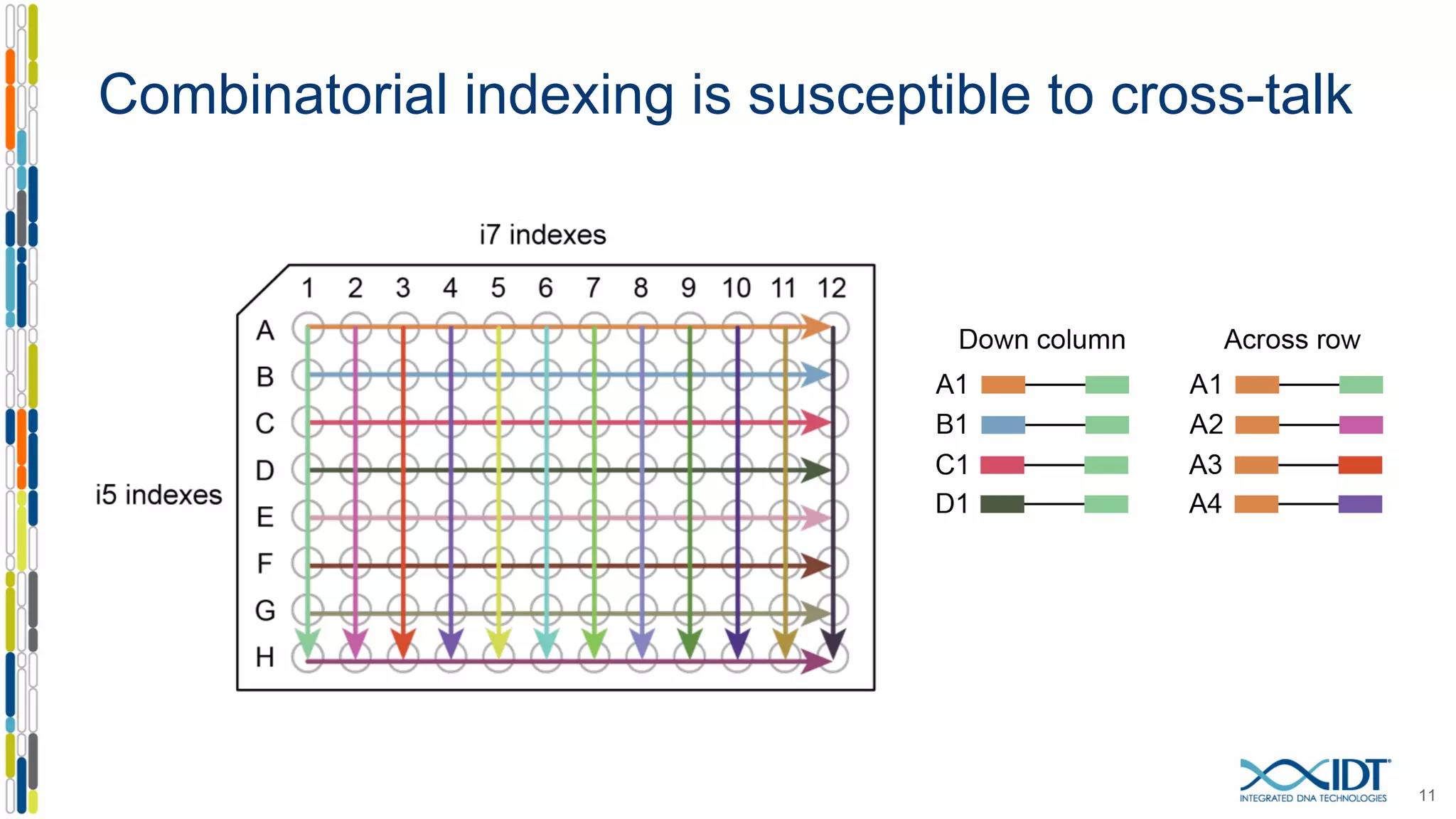
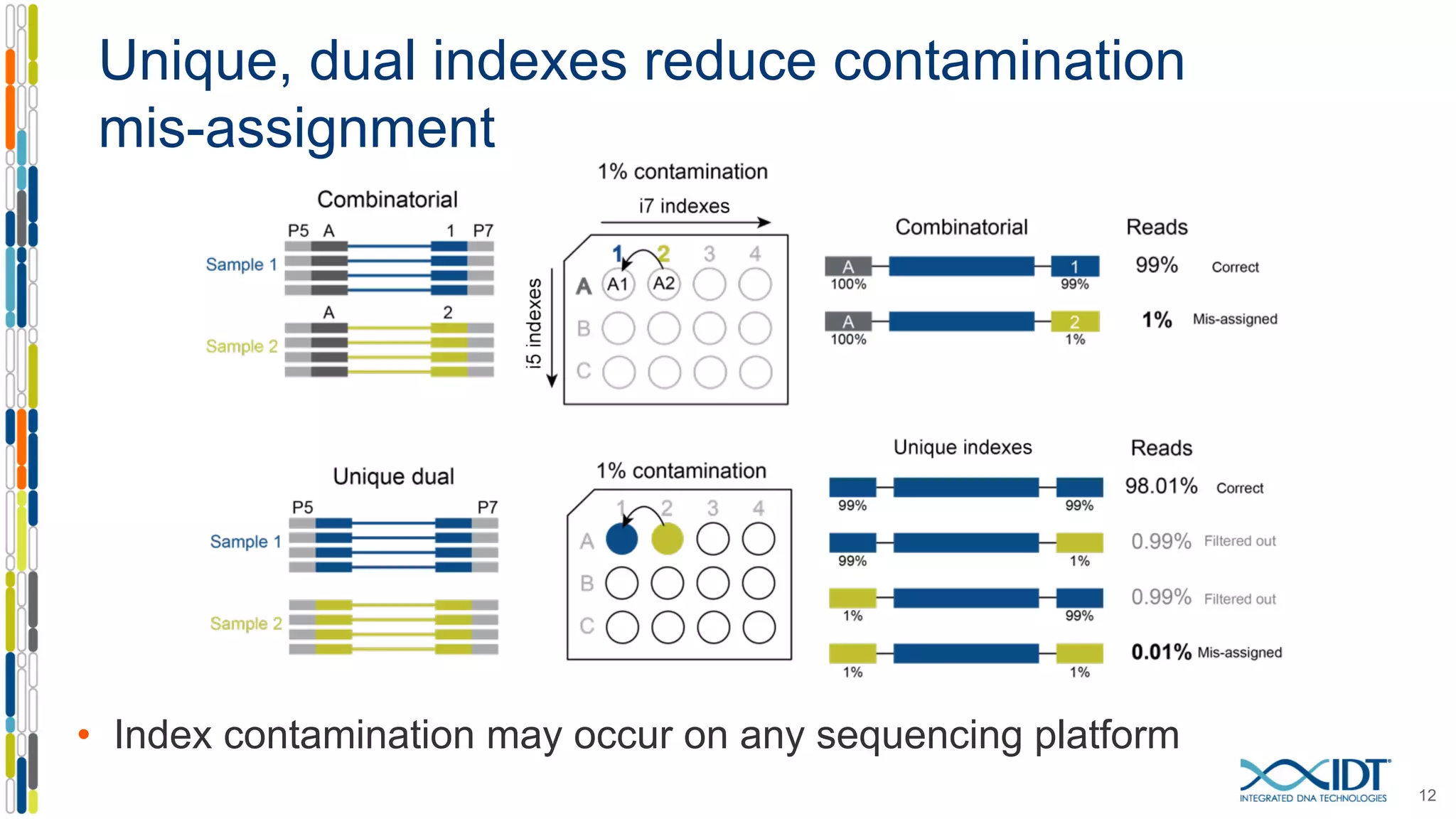
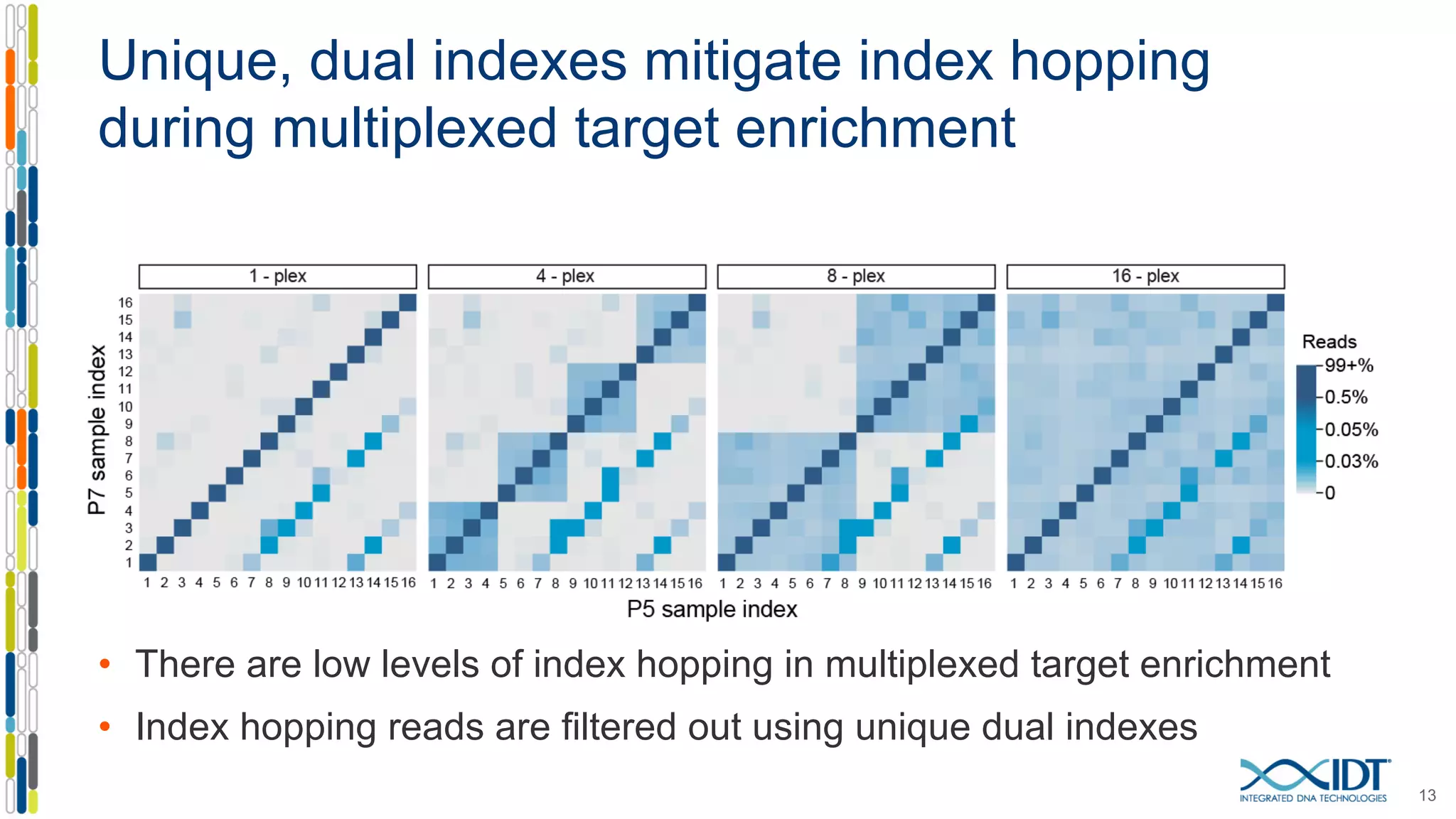
The document presents the Xgen® dual index UMI adapters designed for Illumina sequencers, detailing their role in reducing sample cross-talk and improving the accuracy of low-frequency variant detection in NGS workflows. It discusses various sources of sample cross-talk and how unique, dual indexes can mitigate index hopping and contamination issues. The effectiveness of these adapters is highlighted through consensus analysis that enhances variant calling accuracy, significantly lowering false positives and increasing positive predictive values.
![Index hopping during multiplexed sequencing
• Patterned flow cells use
exclusion amplification (ExAmp)
chemistry
• ExAmp is associated with
higher levels of index hopping
than bridge amplification
• PCR-free libraries exhibit higher
levels of index hopping than
amplified libraries
14
Sinha R, Stanley G, et al. (2017) Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000
DNA sequencing. bioRxiv, doi: https://doi.org/10.1101/125724.
Illumina (2017) Minimize index hopping in multiplexed runs. www.illumina.com/science/education/minimizing-index-hopping.html
[accessed November 9, 2017].](https://image.slidesharecdn.com/idt-webinardual-index-umi-adapter-120617-171206141337/75/Dual-index-adapters-with-UMIs-resolve-index-hopping-and-increase-sensitivity-of-variant-detection-14-2048.jpg)