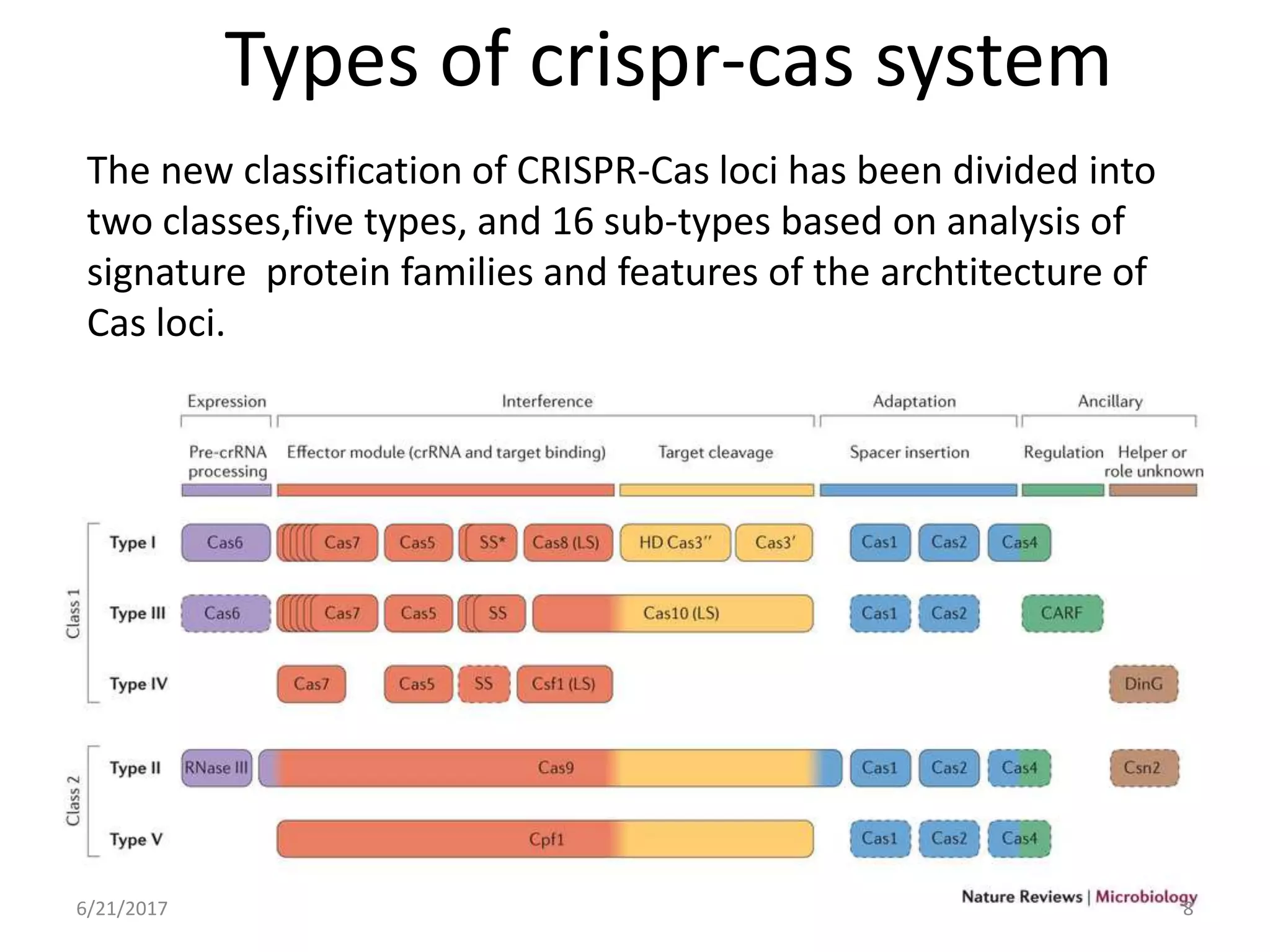
1. CRISPR is a bacterial immune system that provides immunity against viruses. It works by matching DNA sequences from viruses to DNA spacers and cleaving invading DNA.
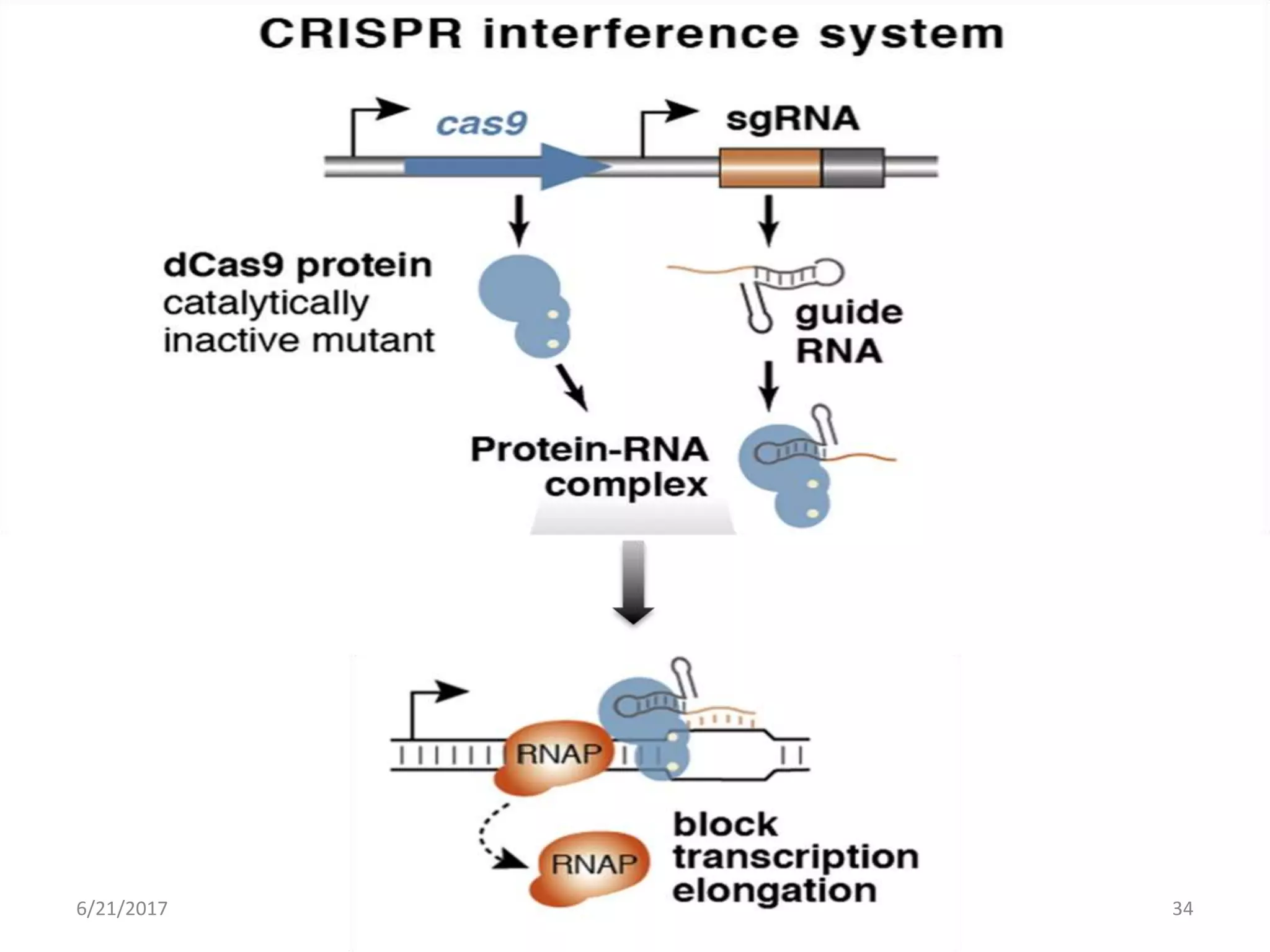
2. The type II CRISPR system is the most well studied and requires only three components - Cas9 protein, CRISPR RNA (crRNA), and trans-activating CRISPR RNA (tracrRNA) - to function. Combining crRNA and tracrRNA into a single RNA molecule called sgRNA was crucial for developing the CRISPR technique.
3. CRISPR technology has become a powerful genome editing tool due to its simplicity, high efficiency, low cost, and ease of use. It allows targeted