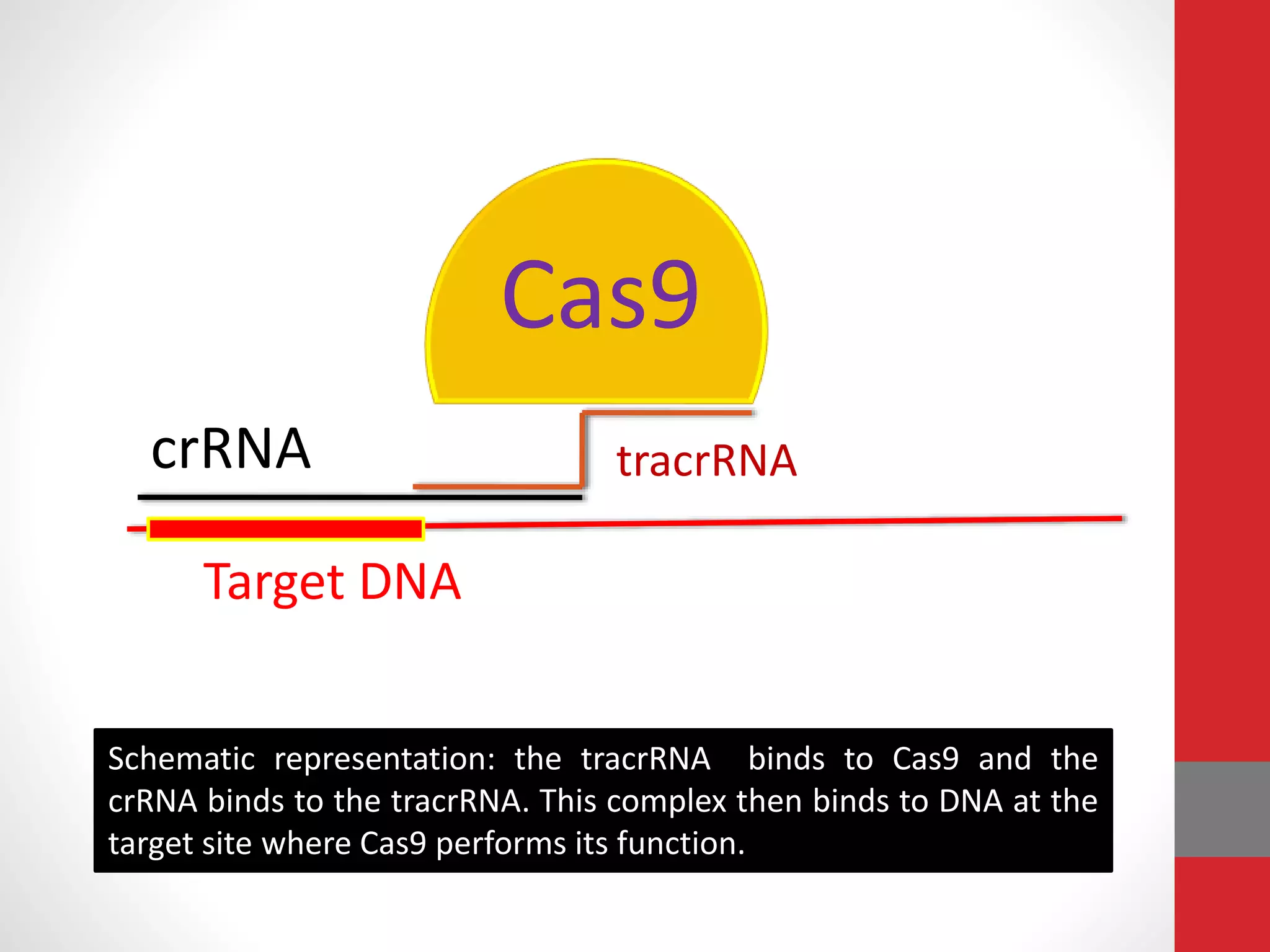
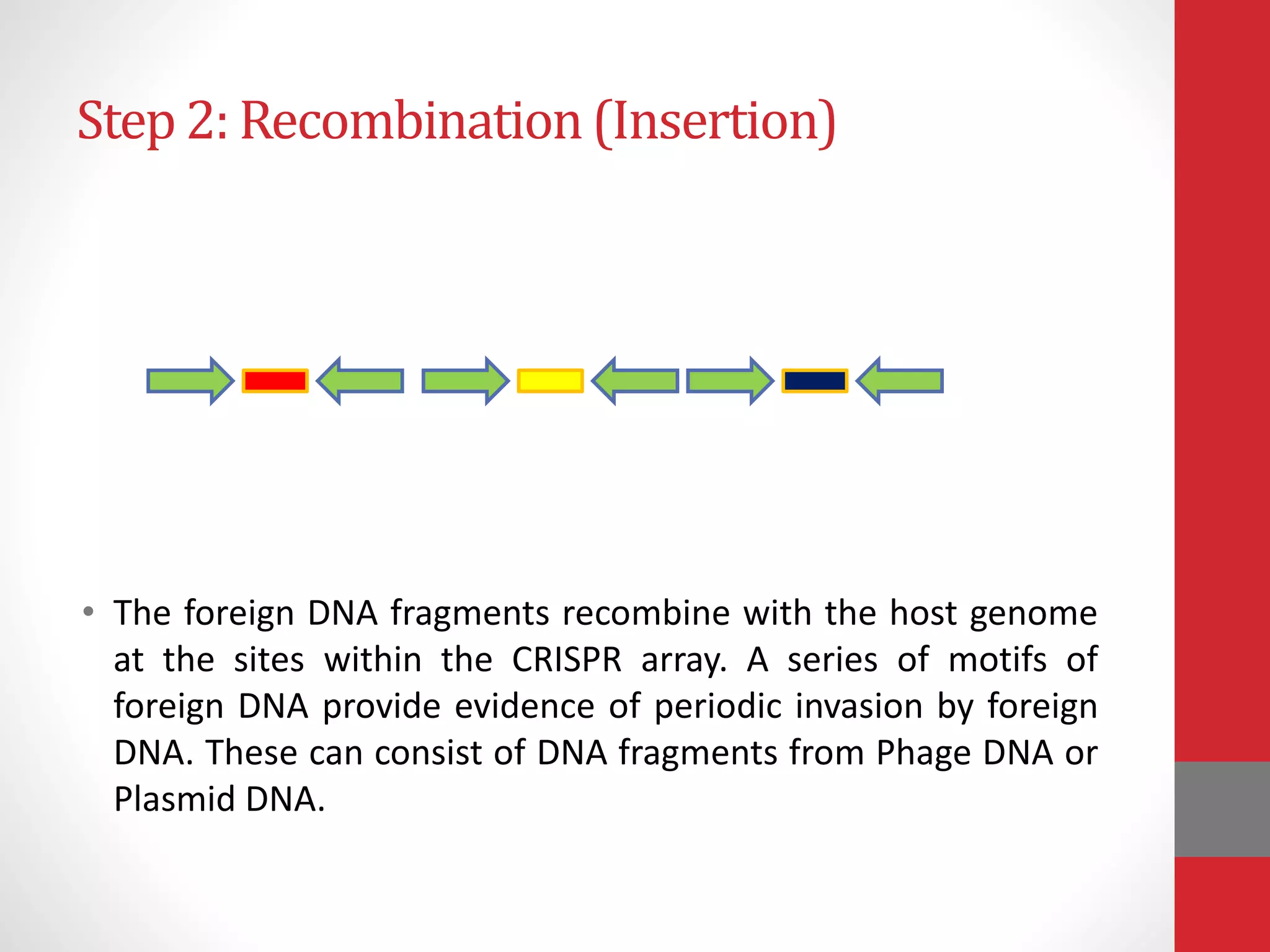
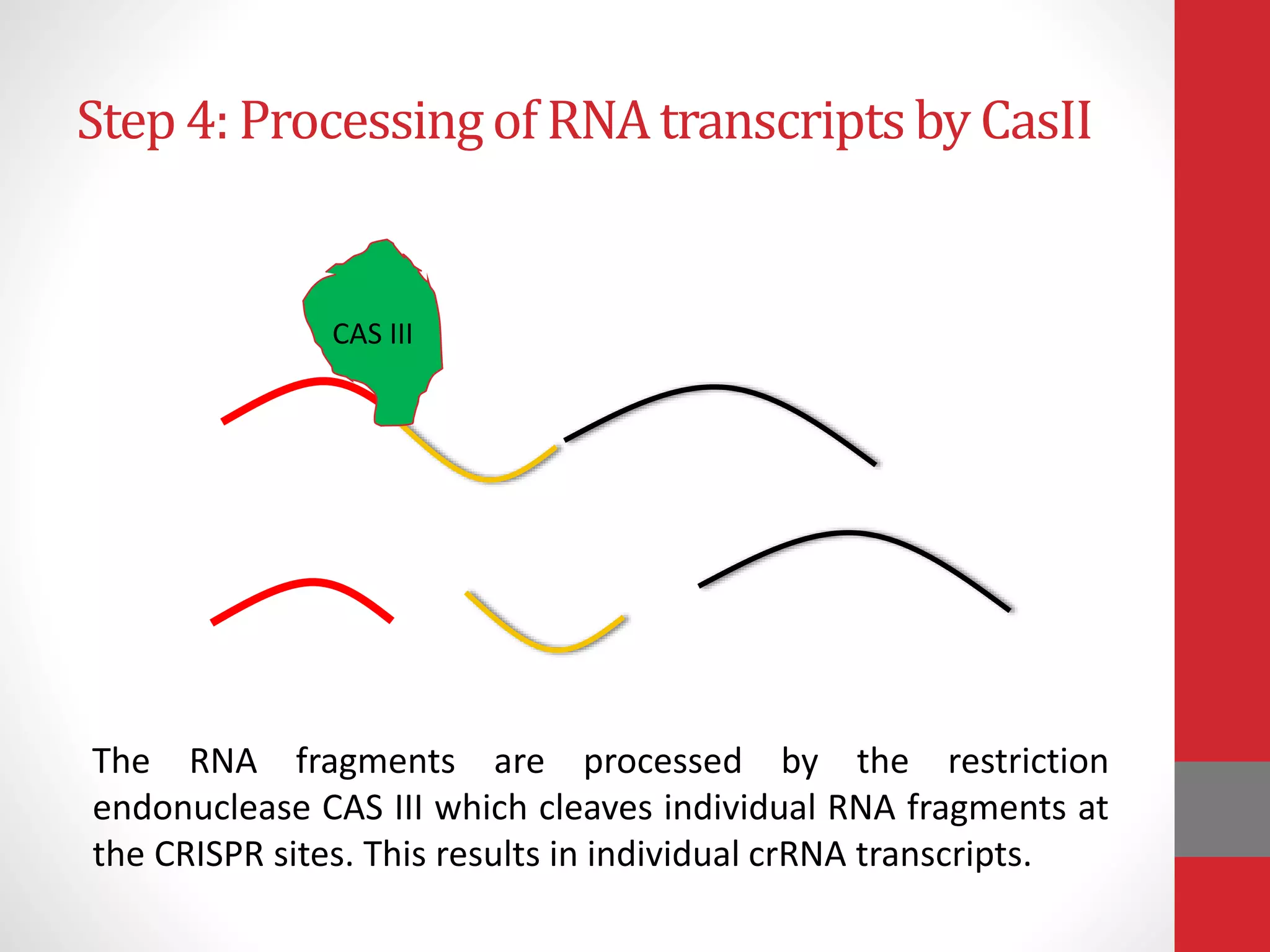
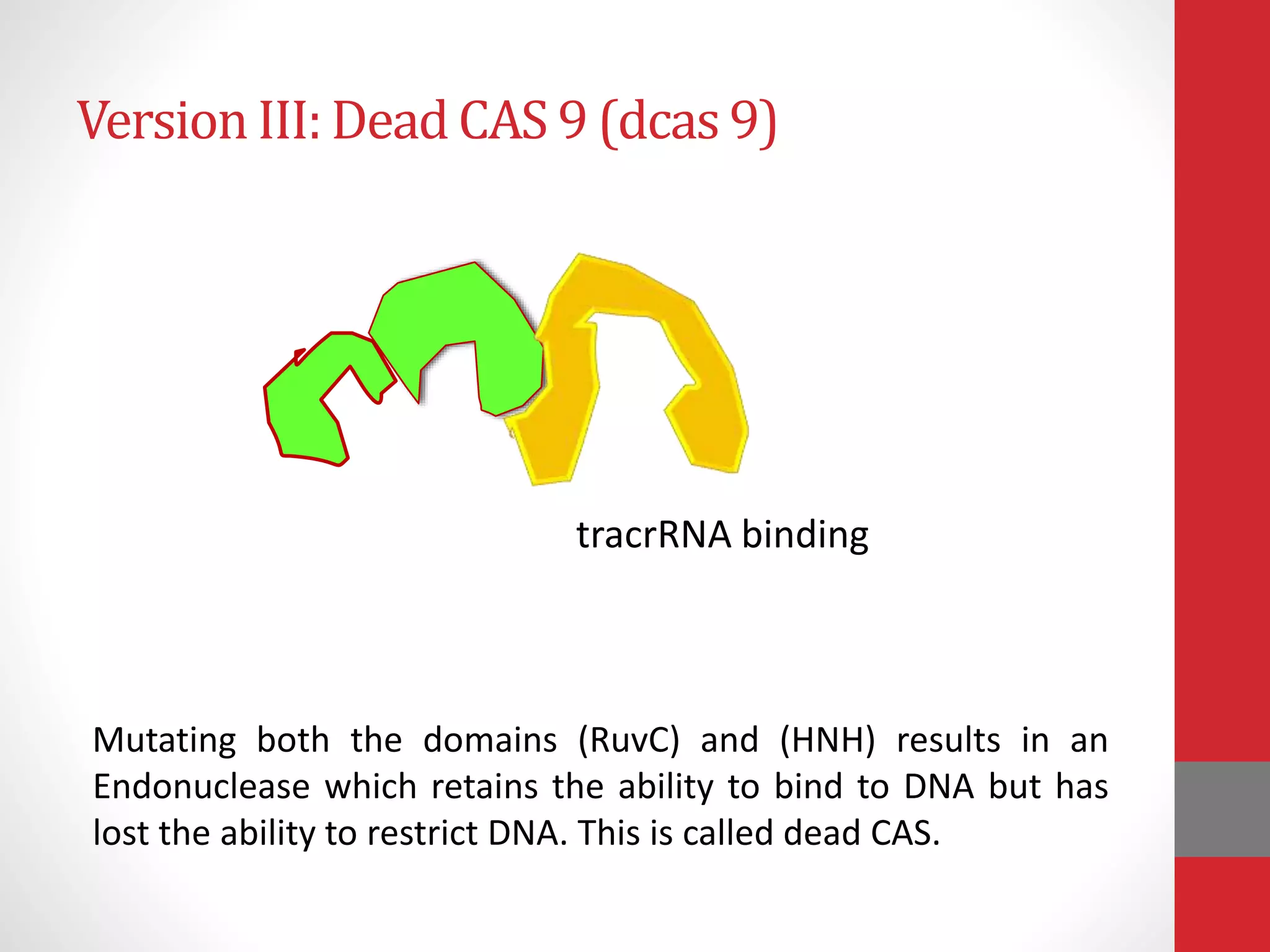
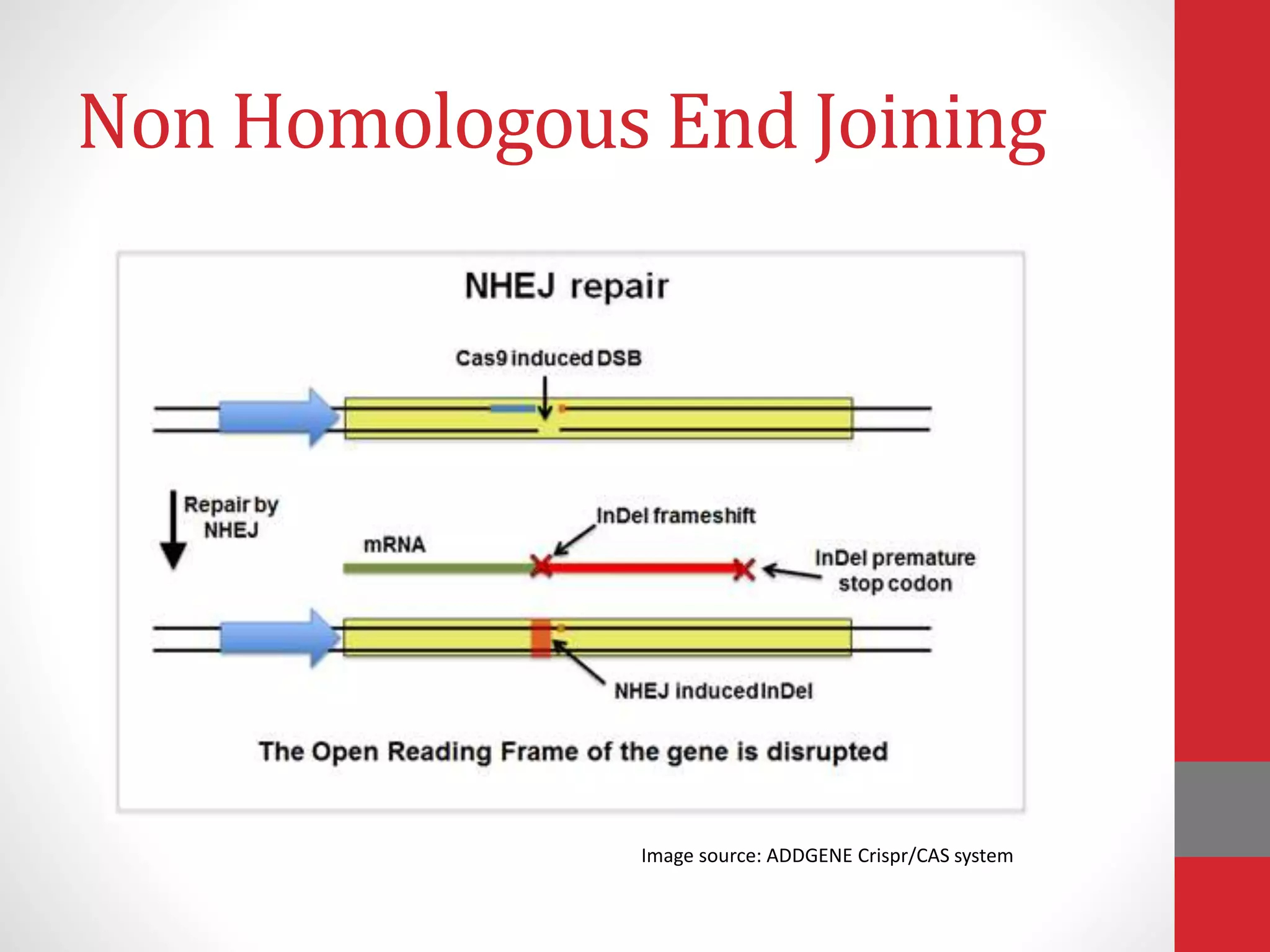
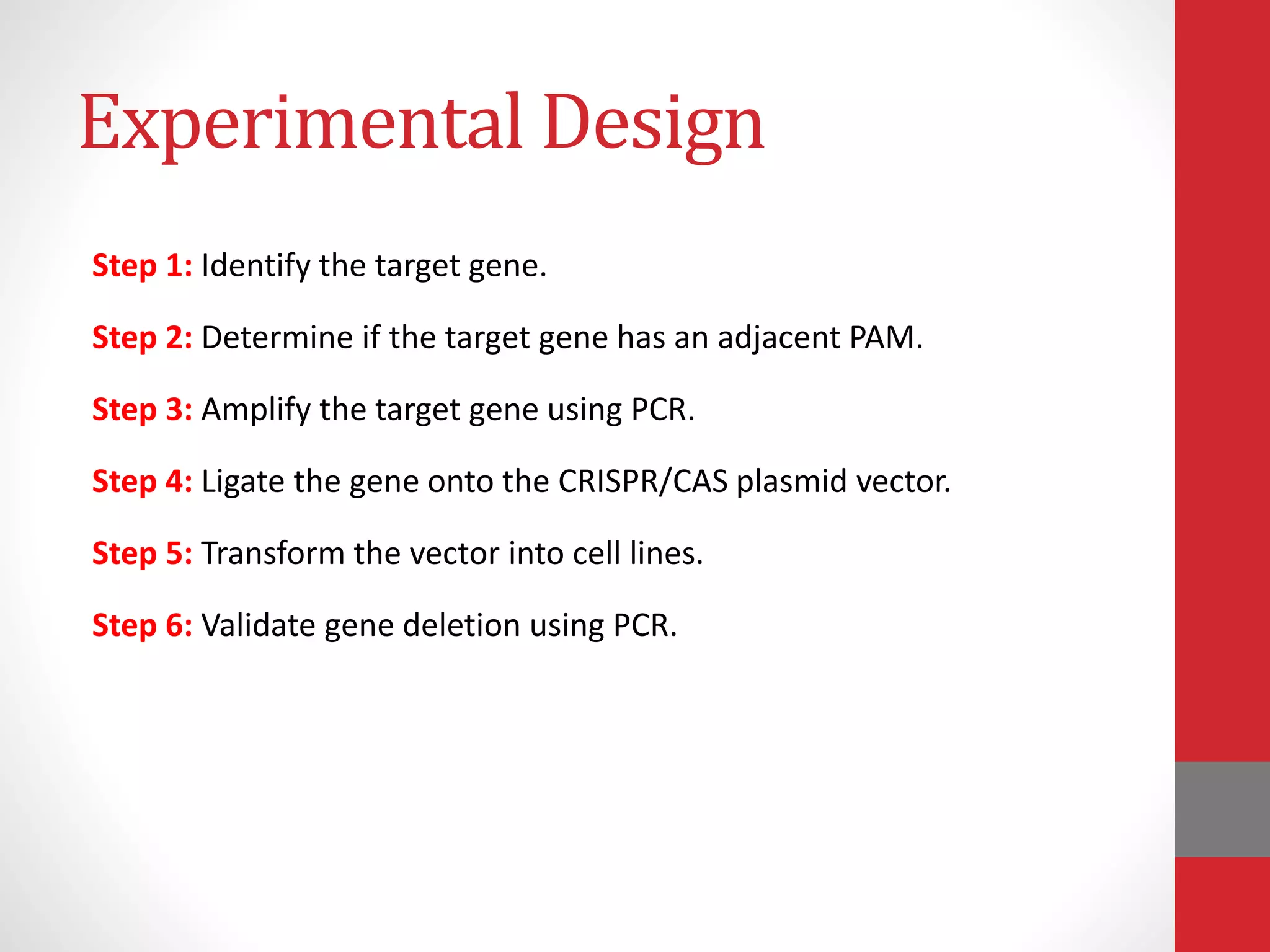
The document provides an overview of the CRISPR/Cas genome editing system, highlighting its components such as Cas9, tracrRNA, and crRNA, as well as its mechanism of action based on bacterial immunity. It outlines objectives and expected learning outcomes related to the system's function in genetic engineering and experimental design for gene deletion. Additionally, it discusses the engineering process for utilizing the CRISPR system in various applications, including the implications for genetically modified organisms.