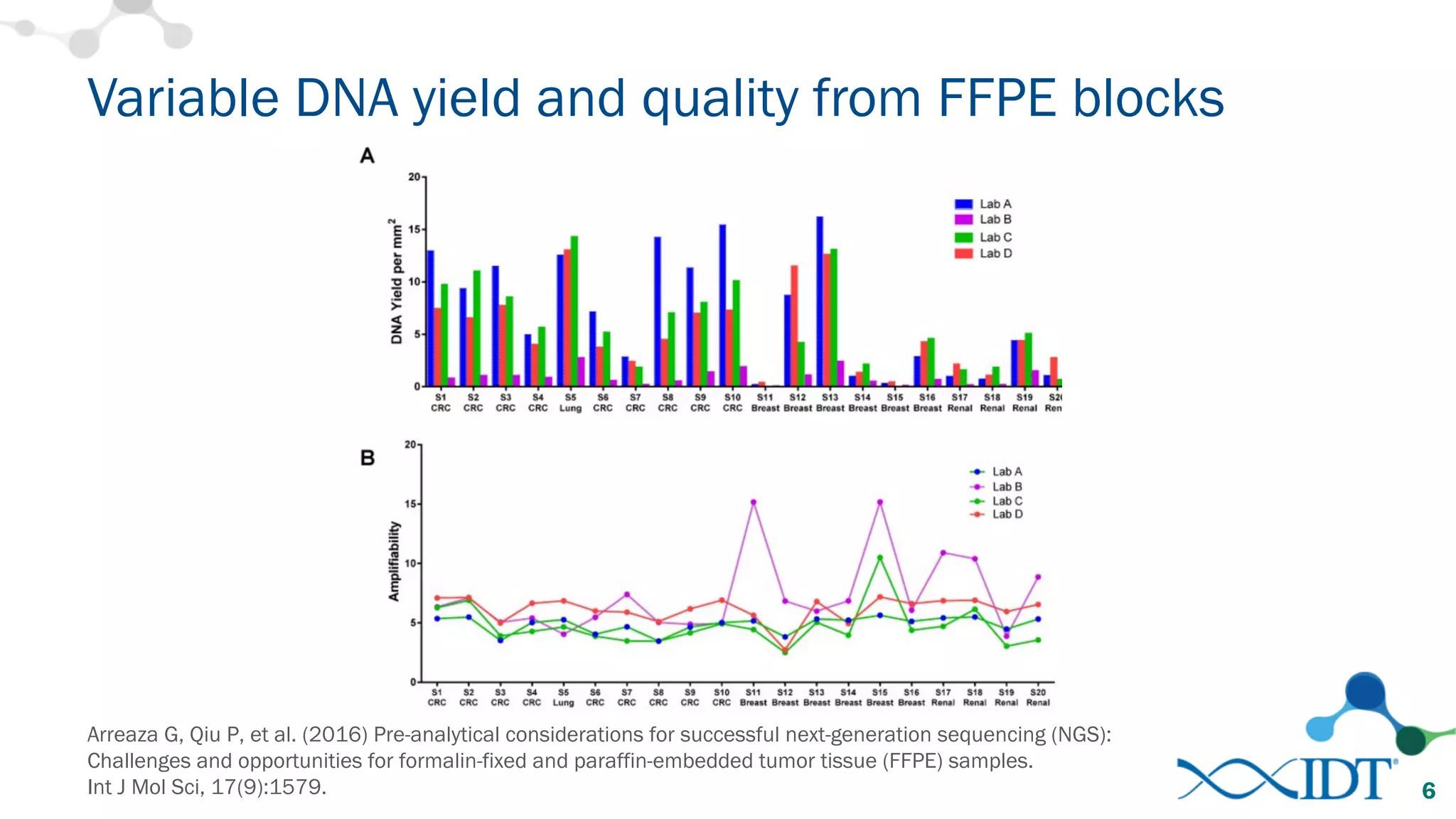
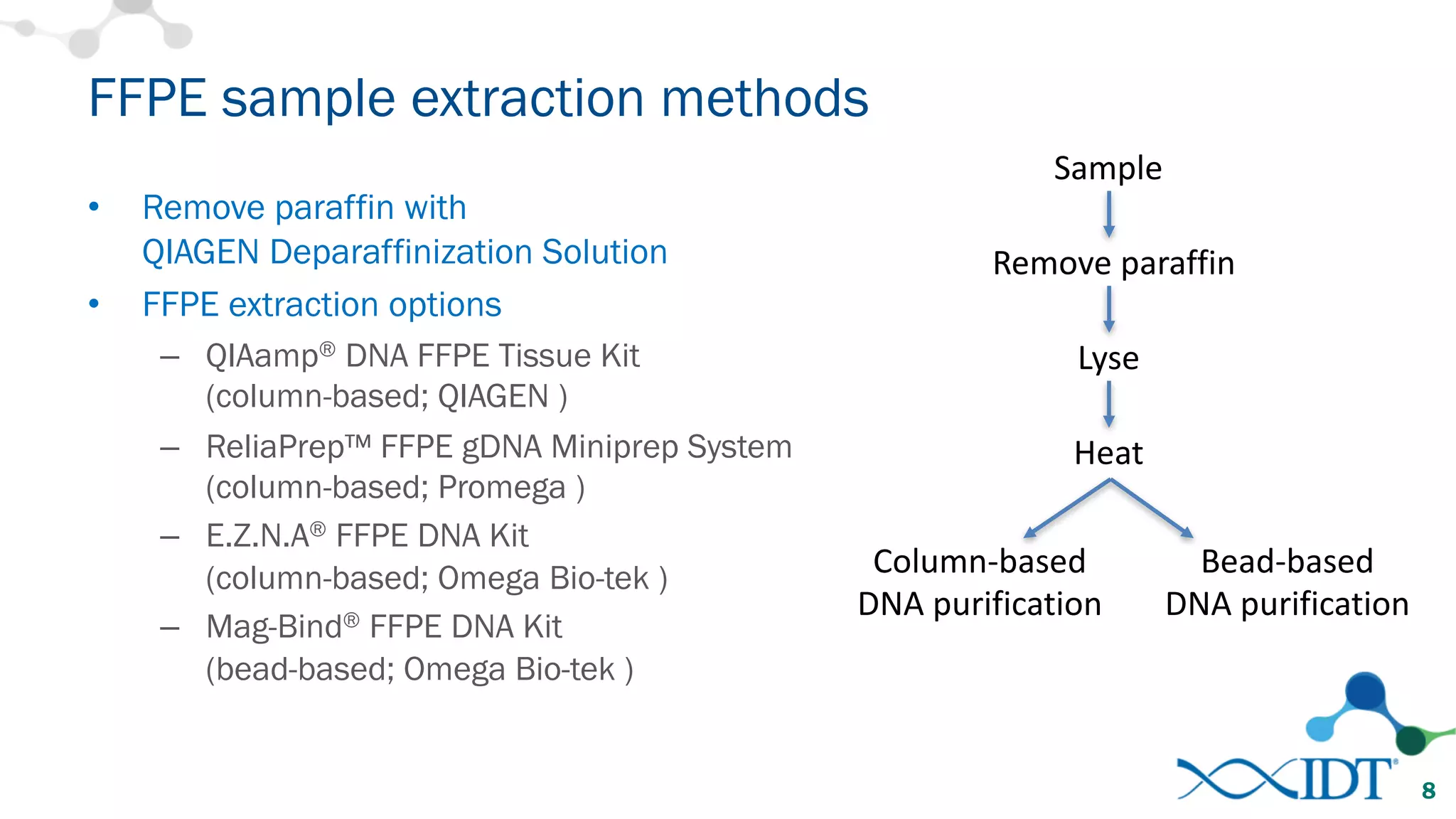
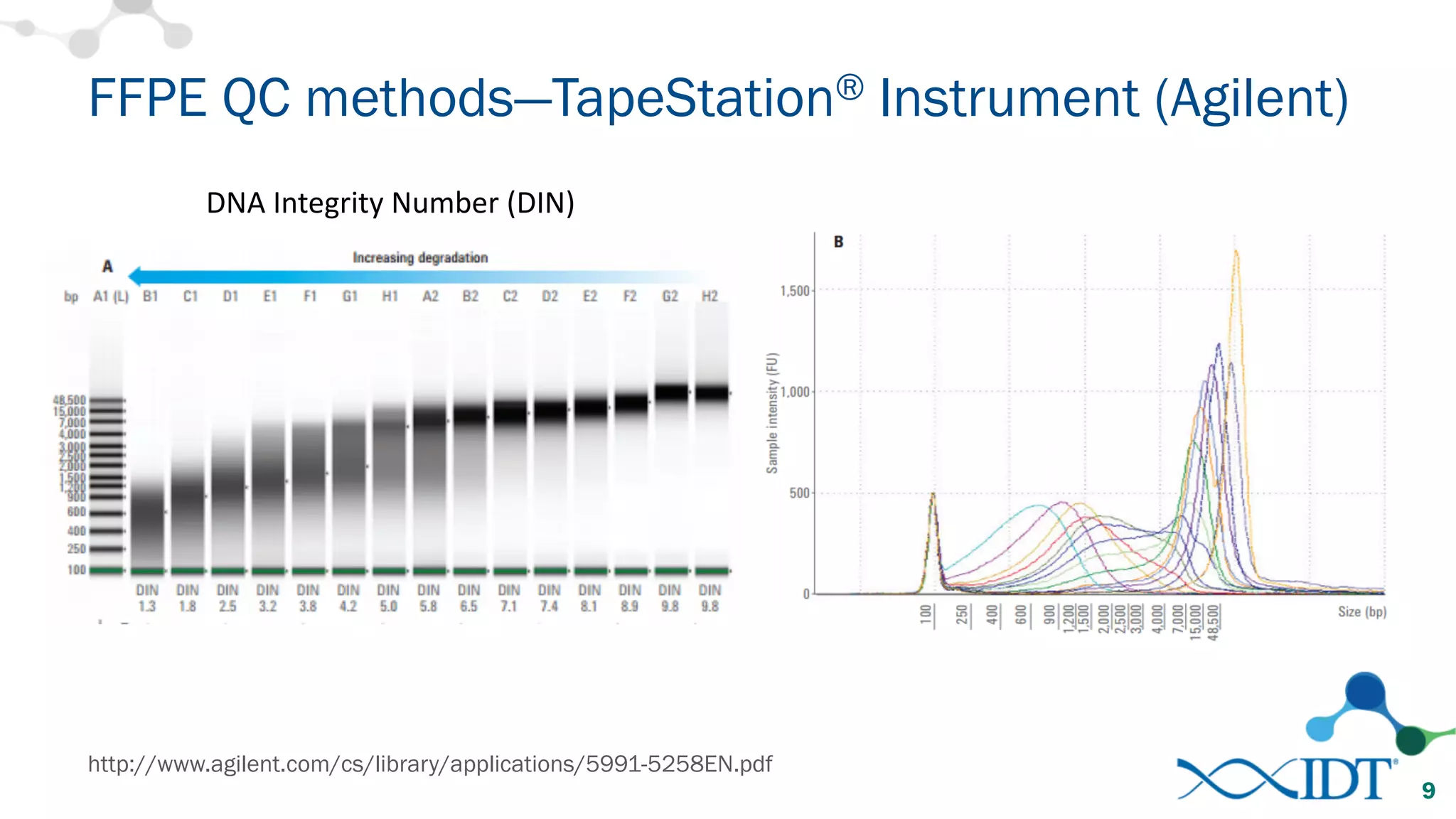
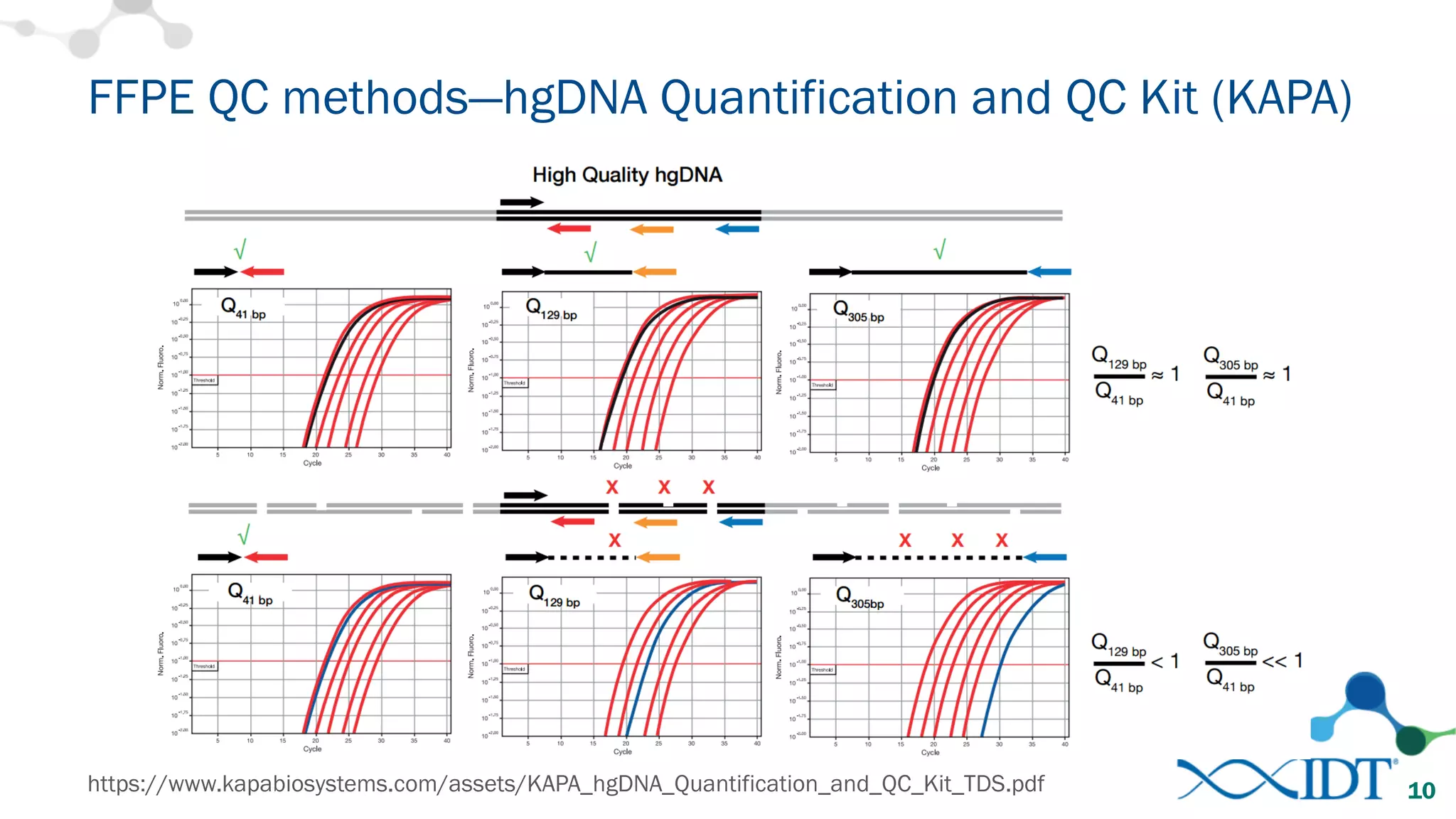
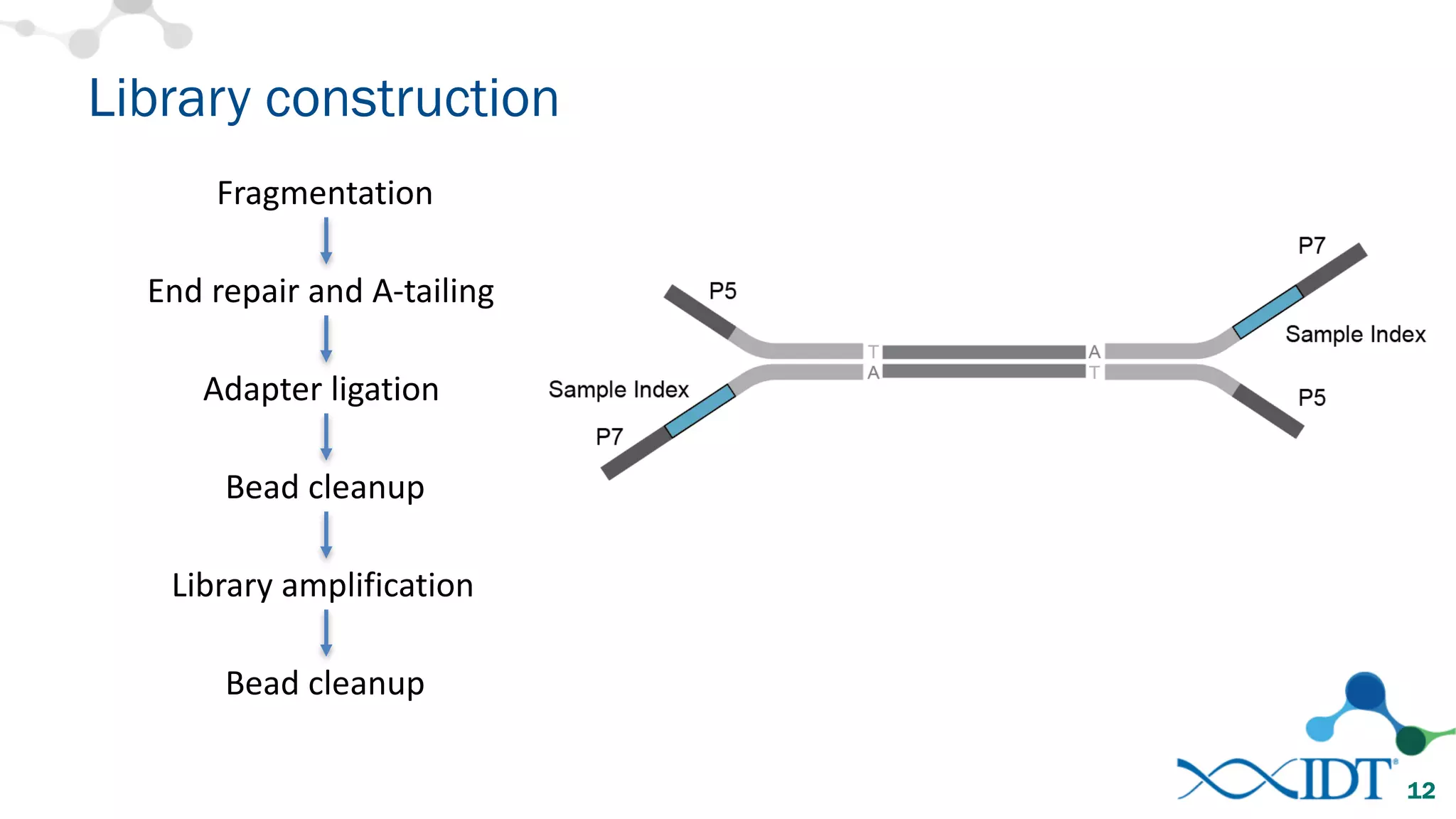
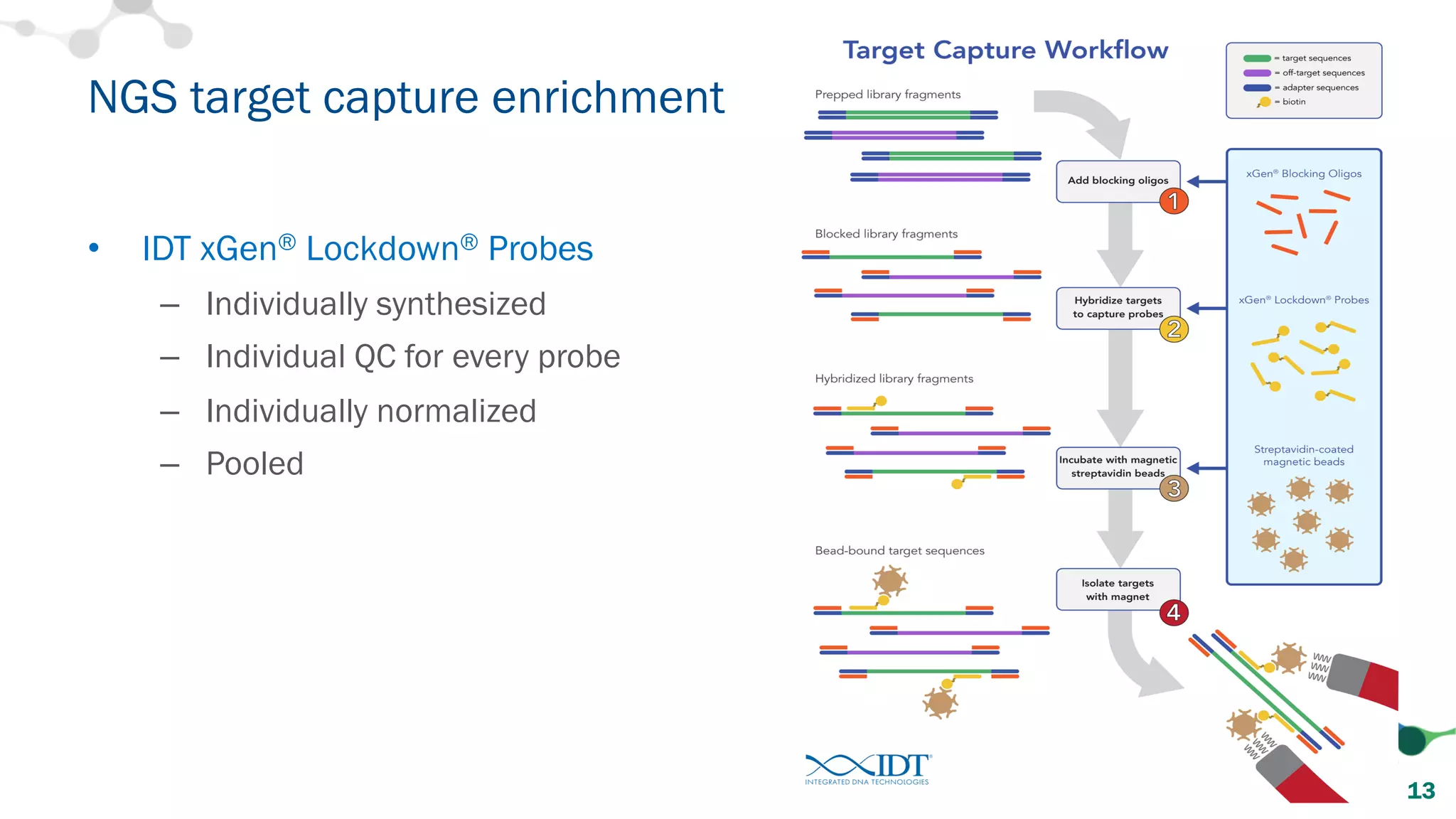
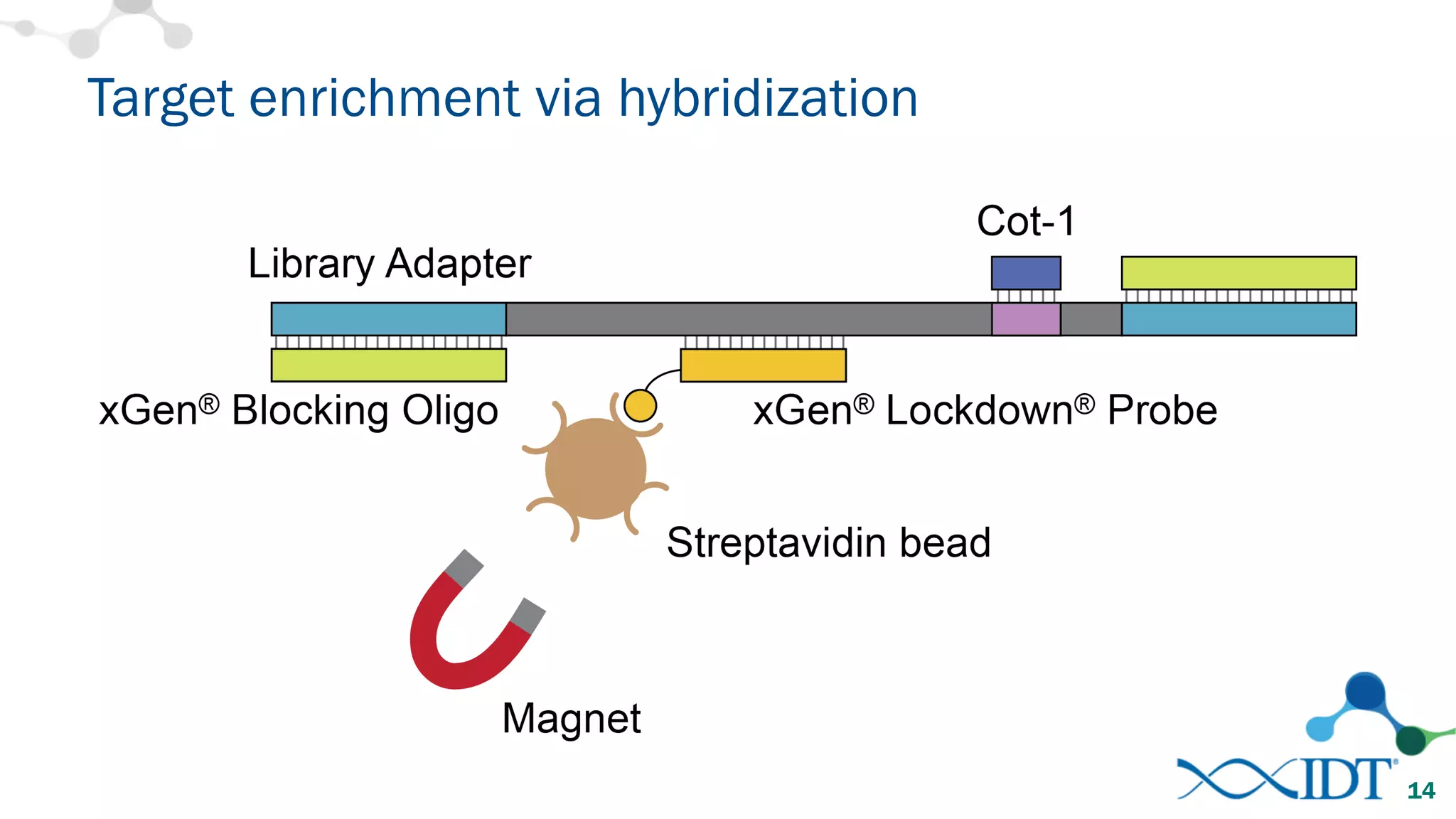
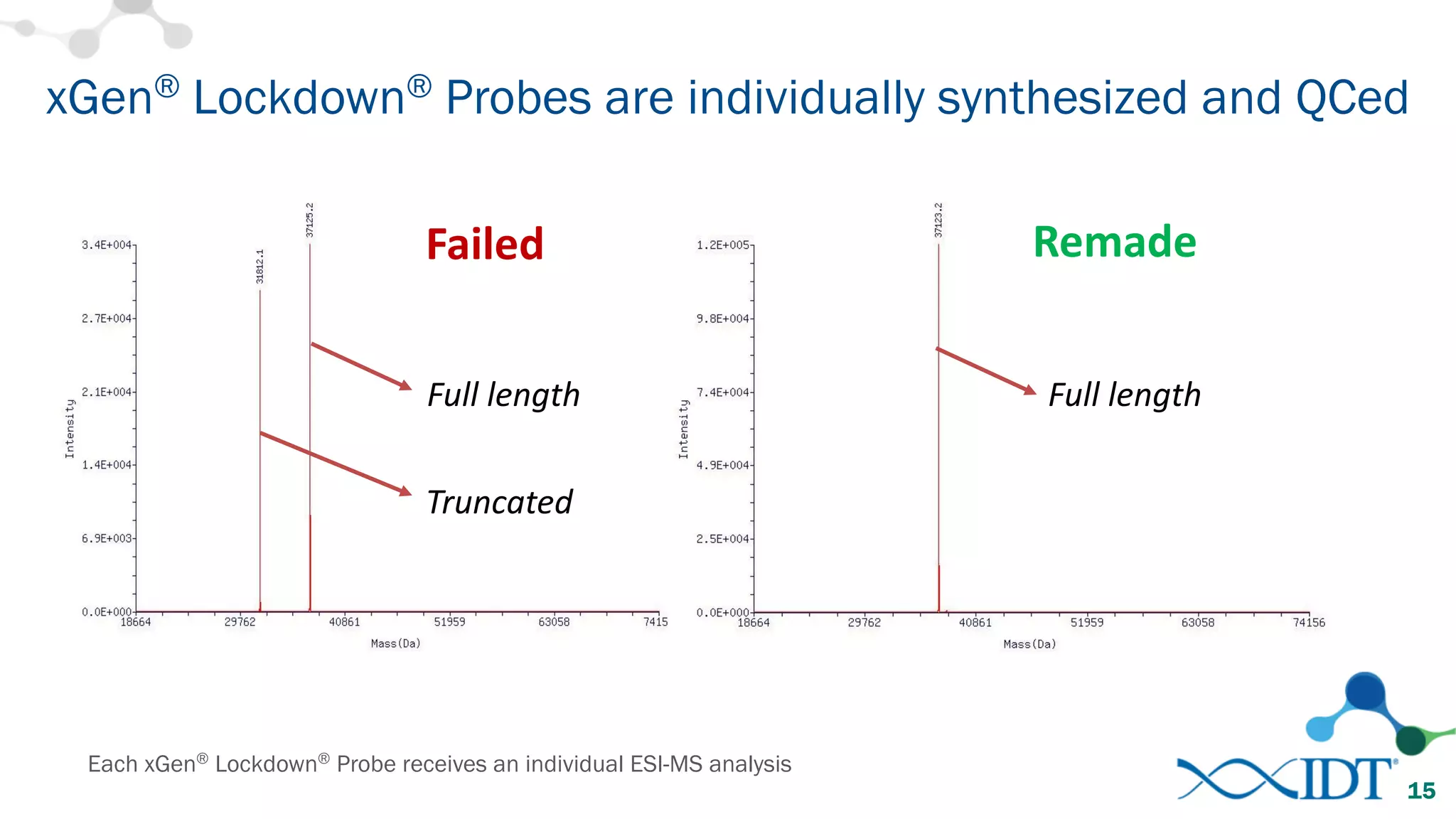
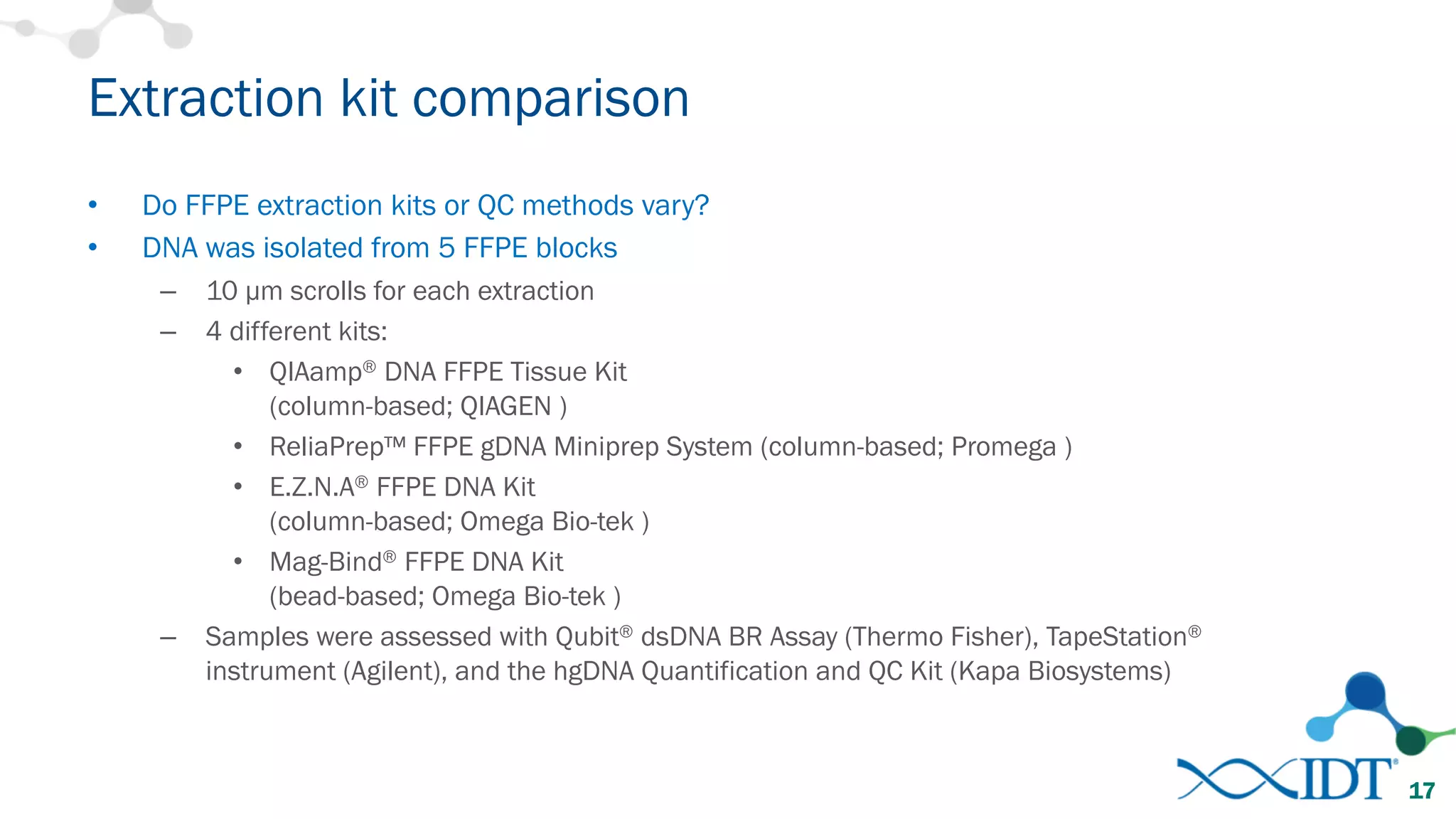
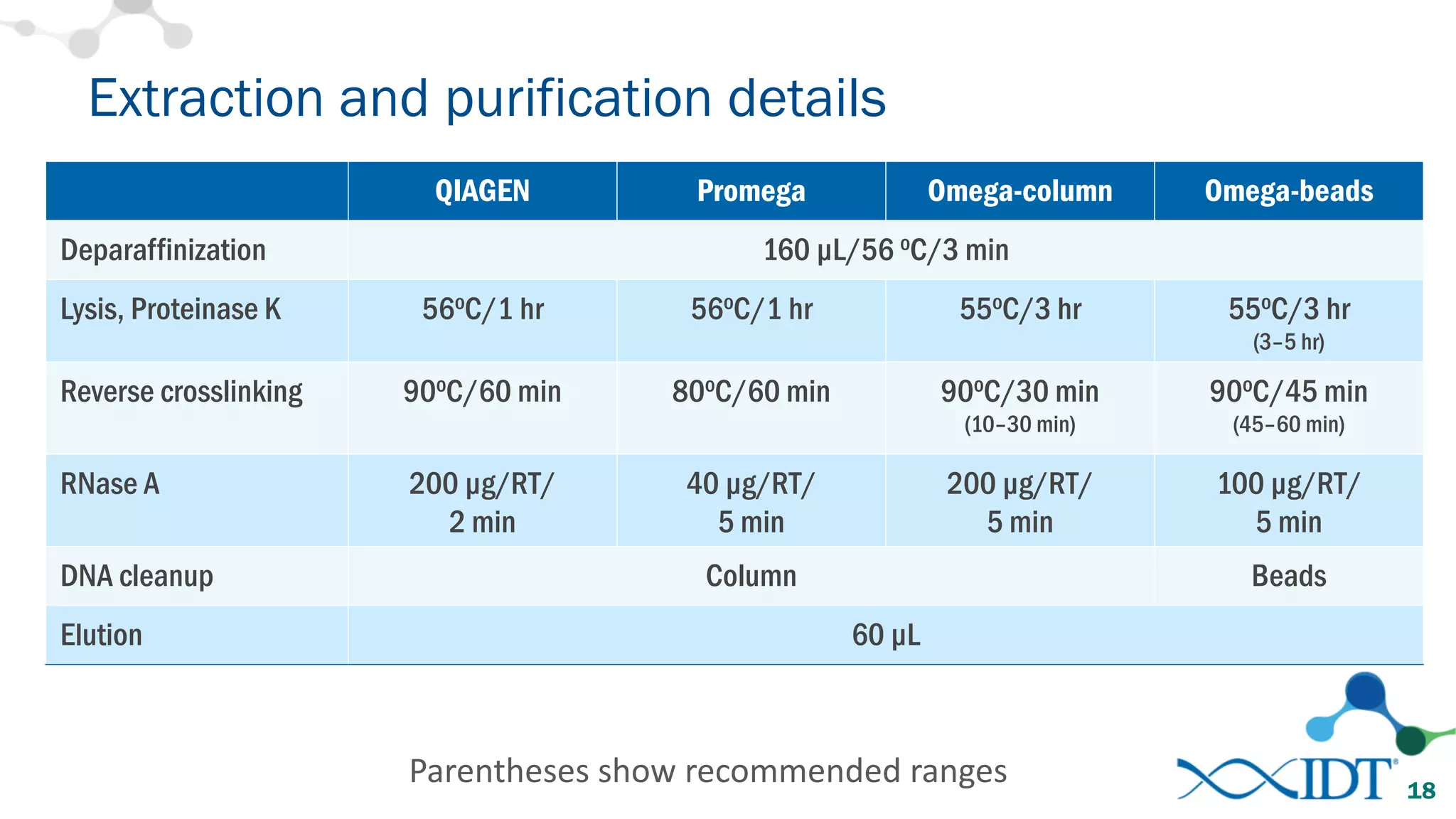
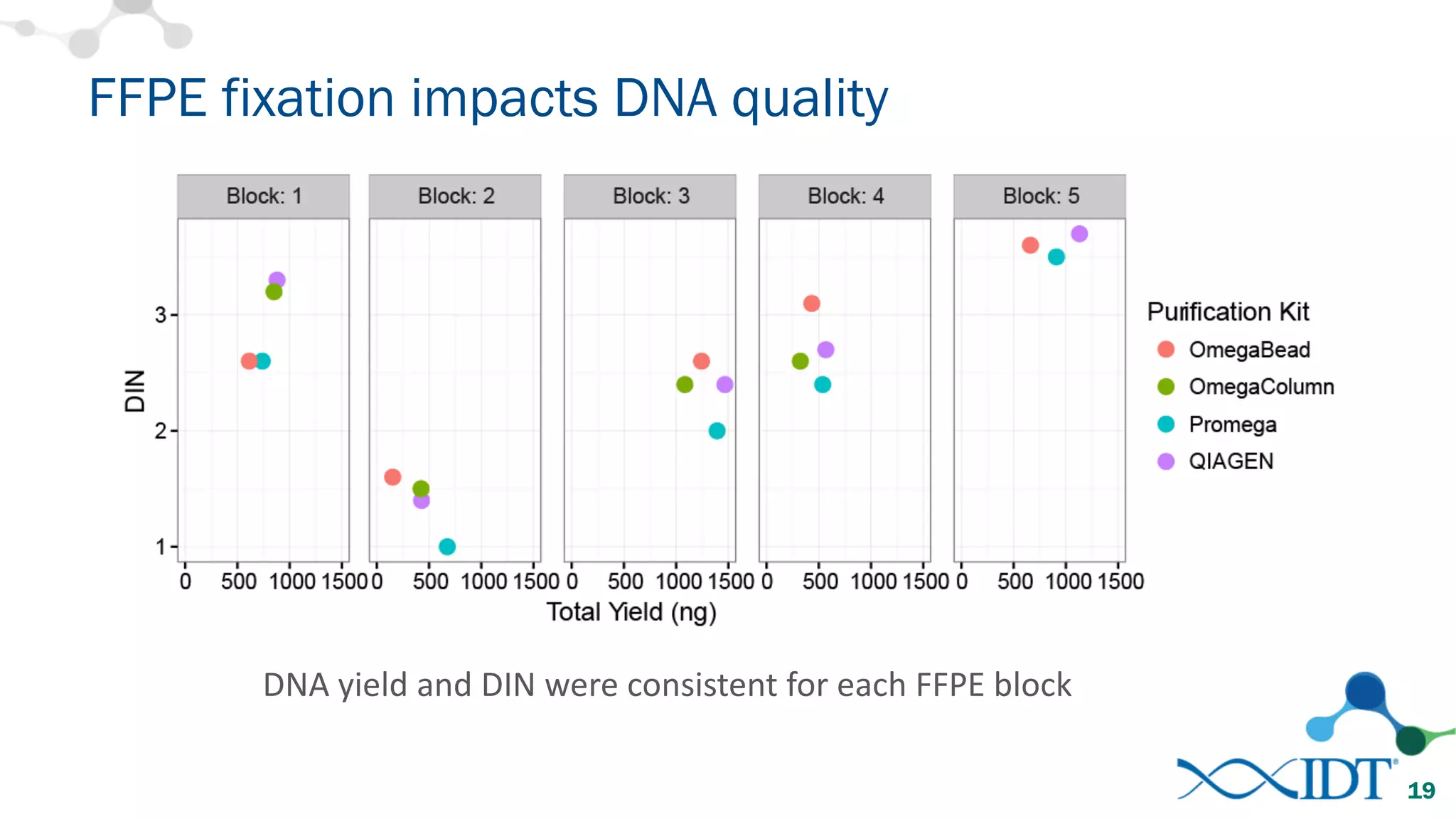
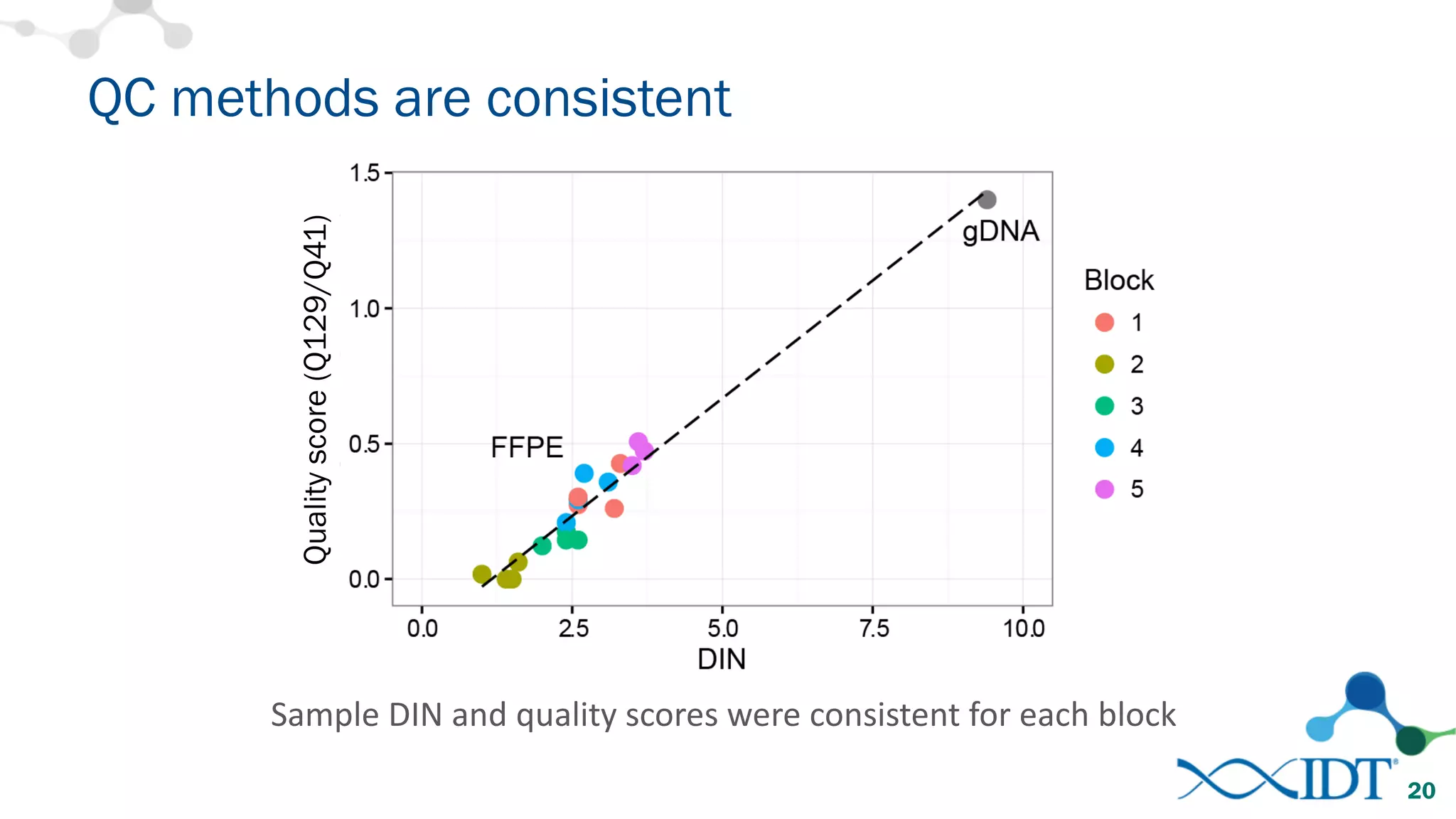
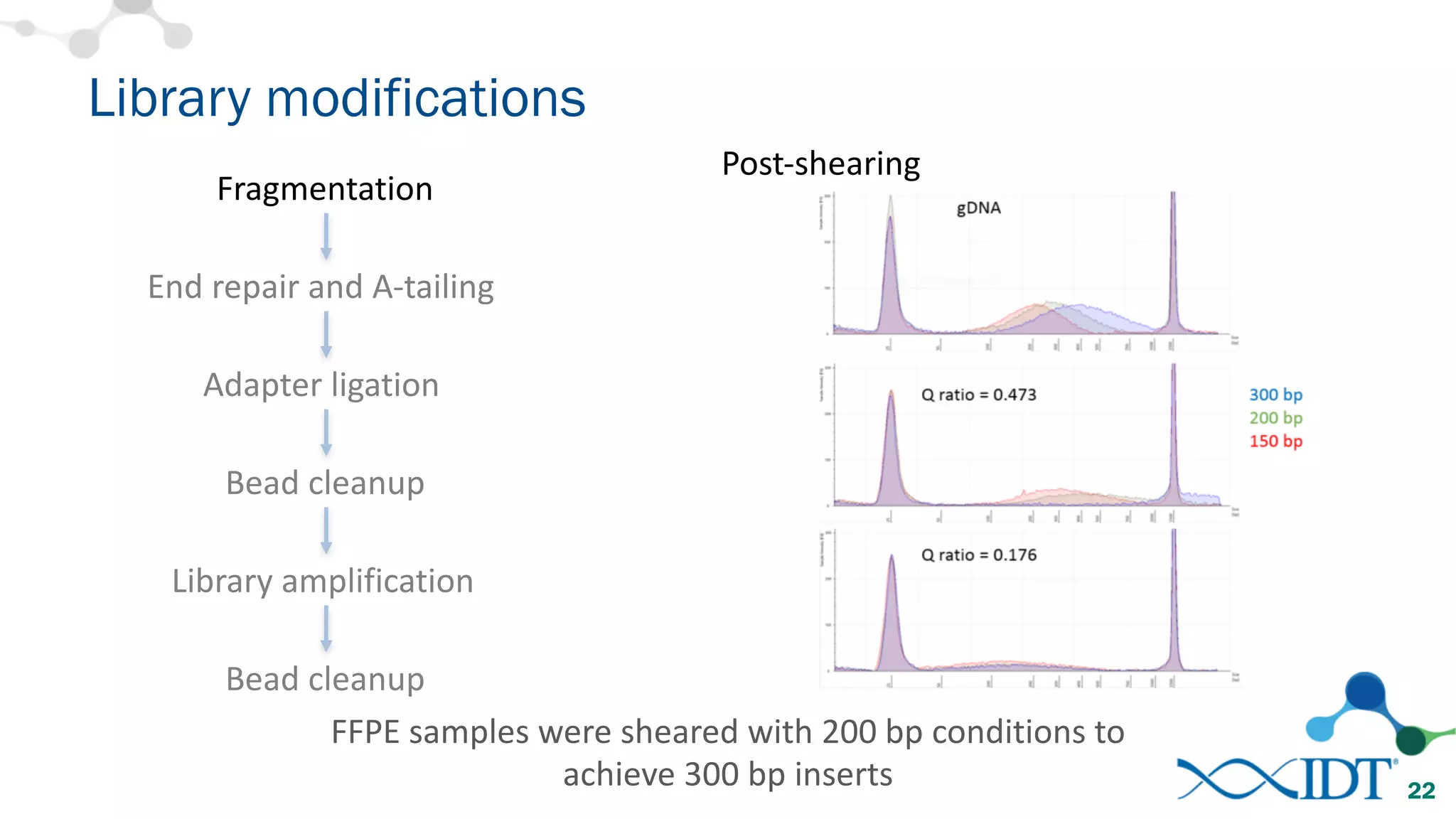
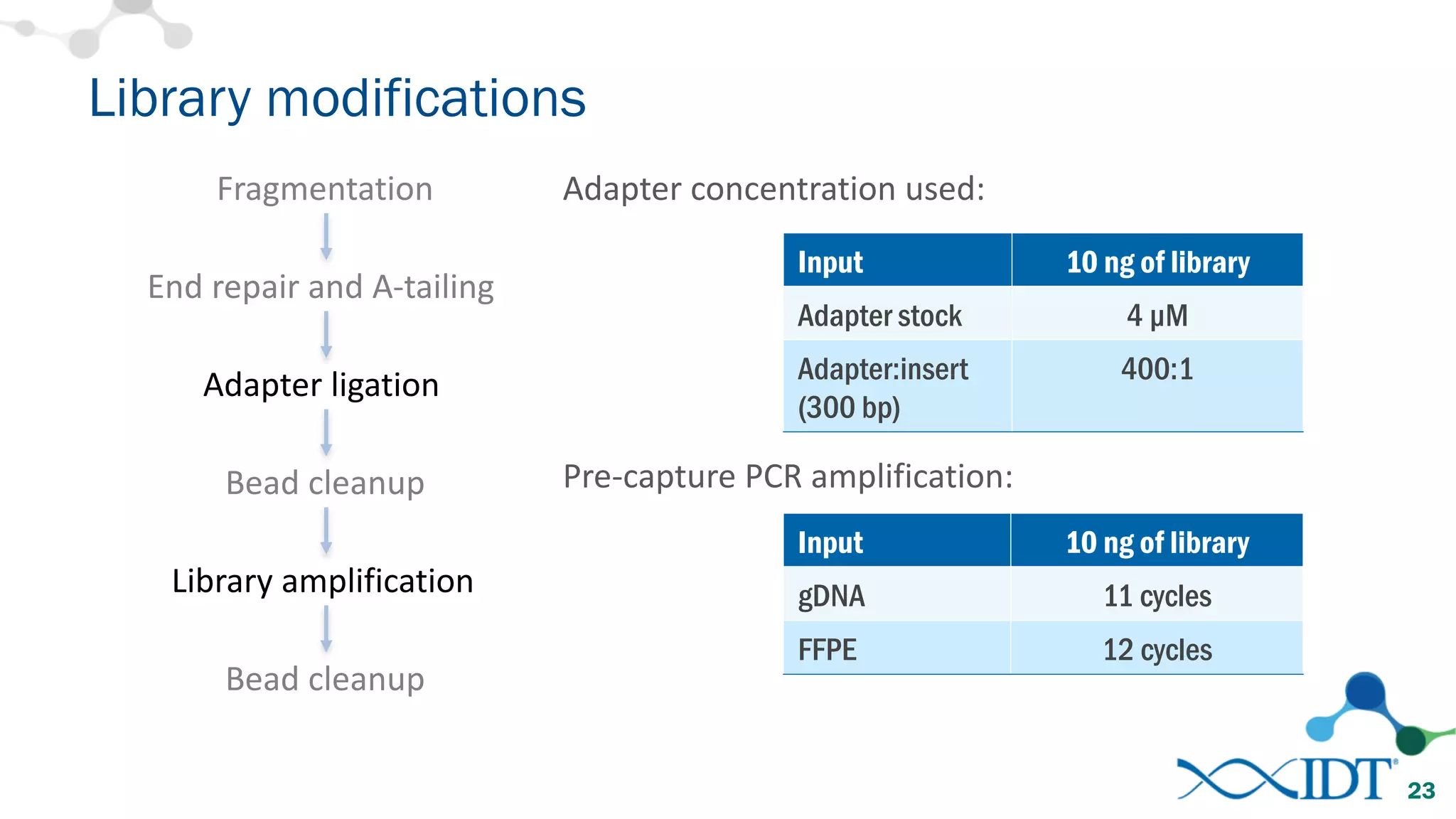
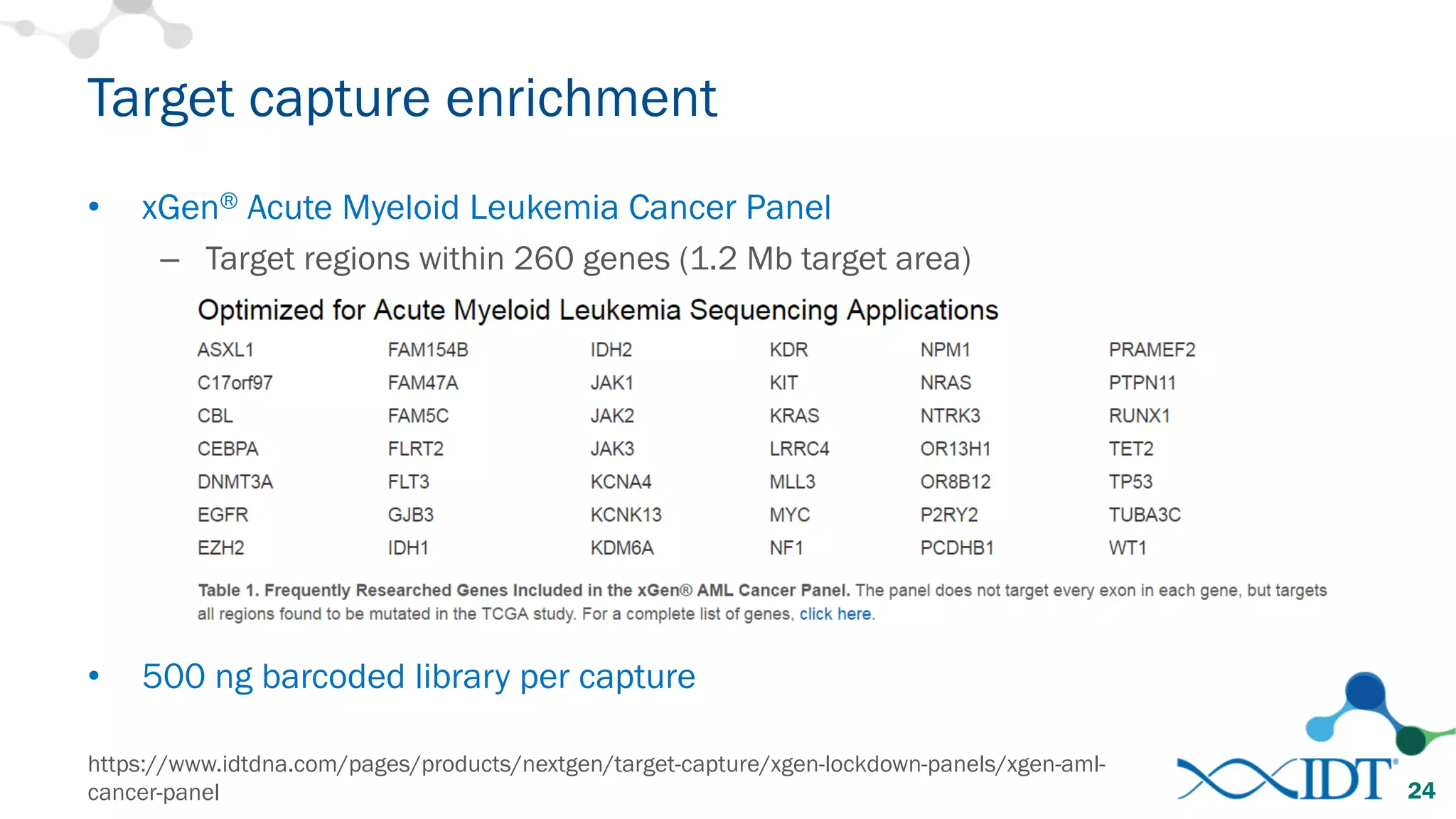
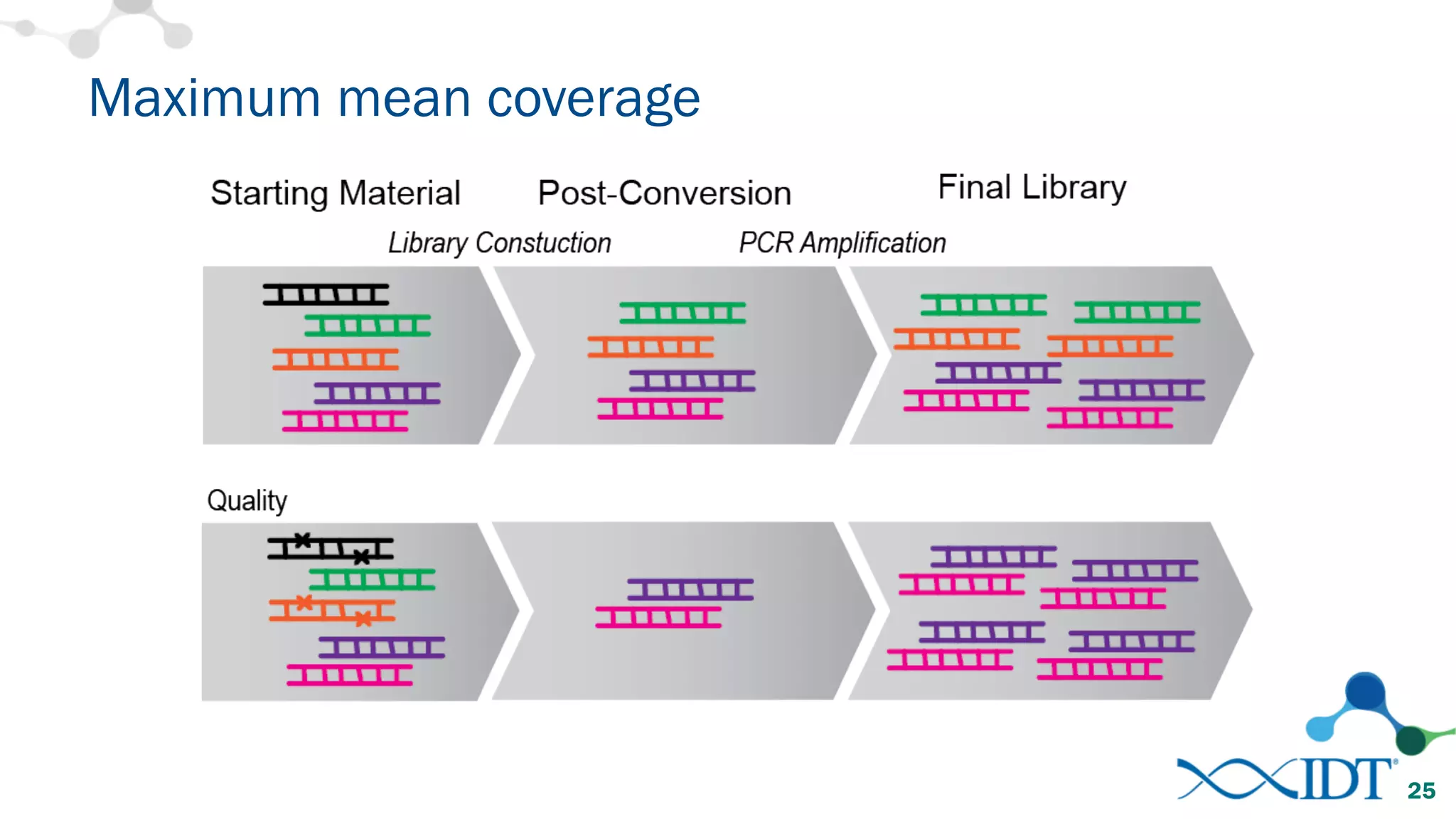
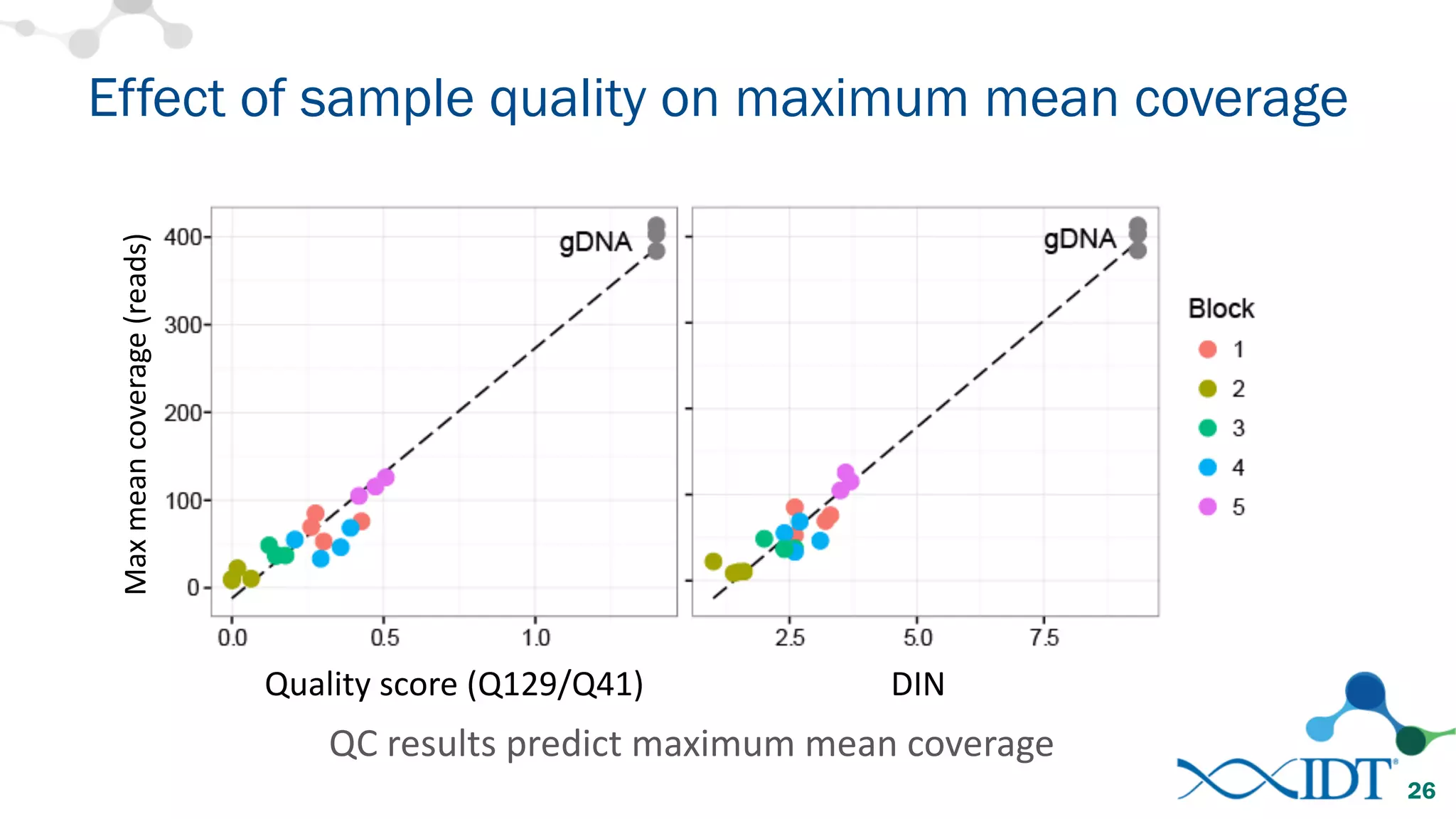
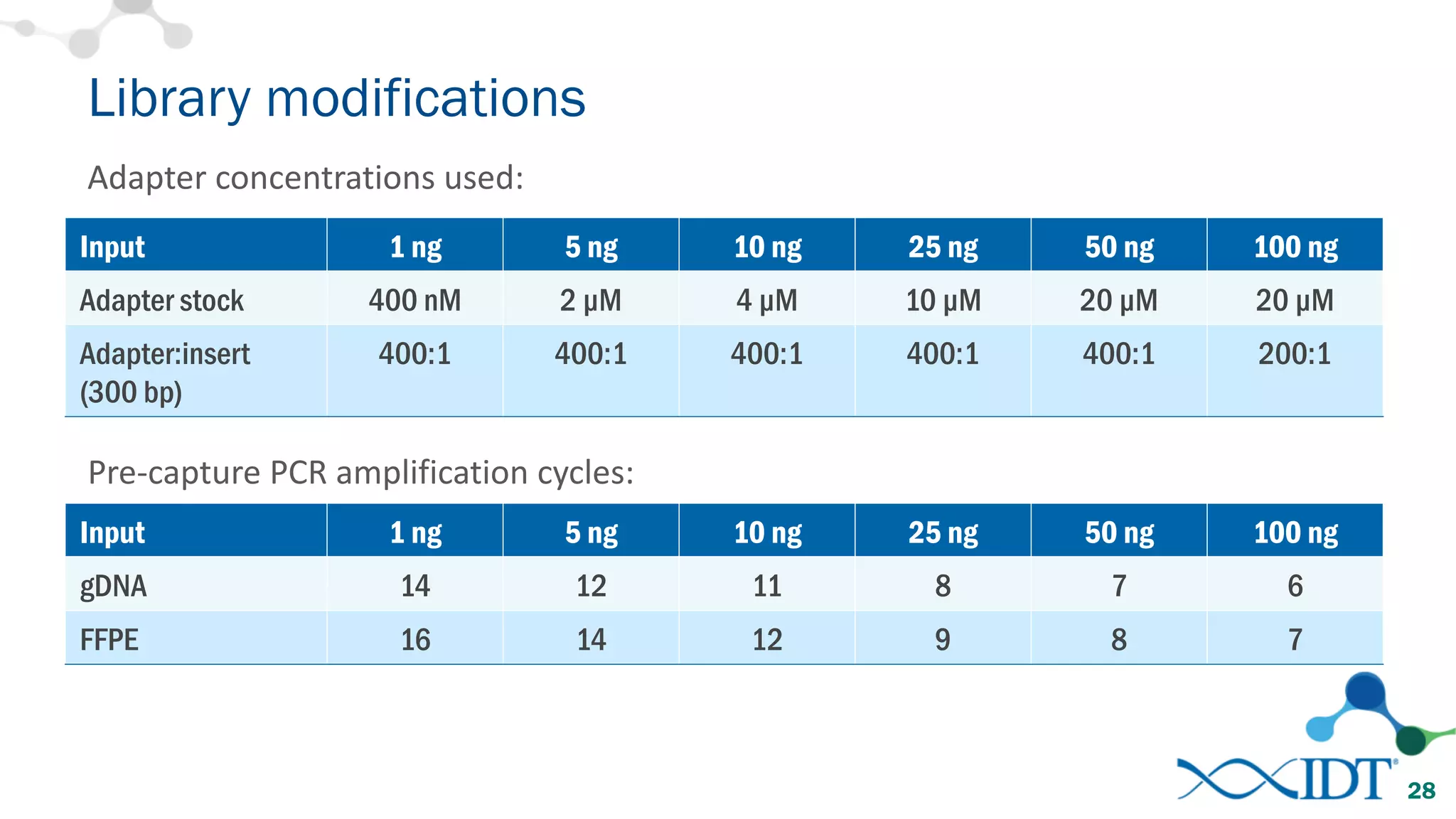
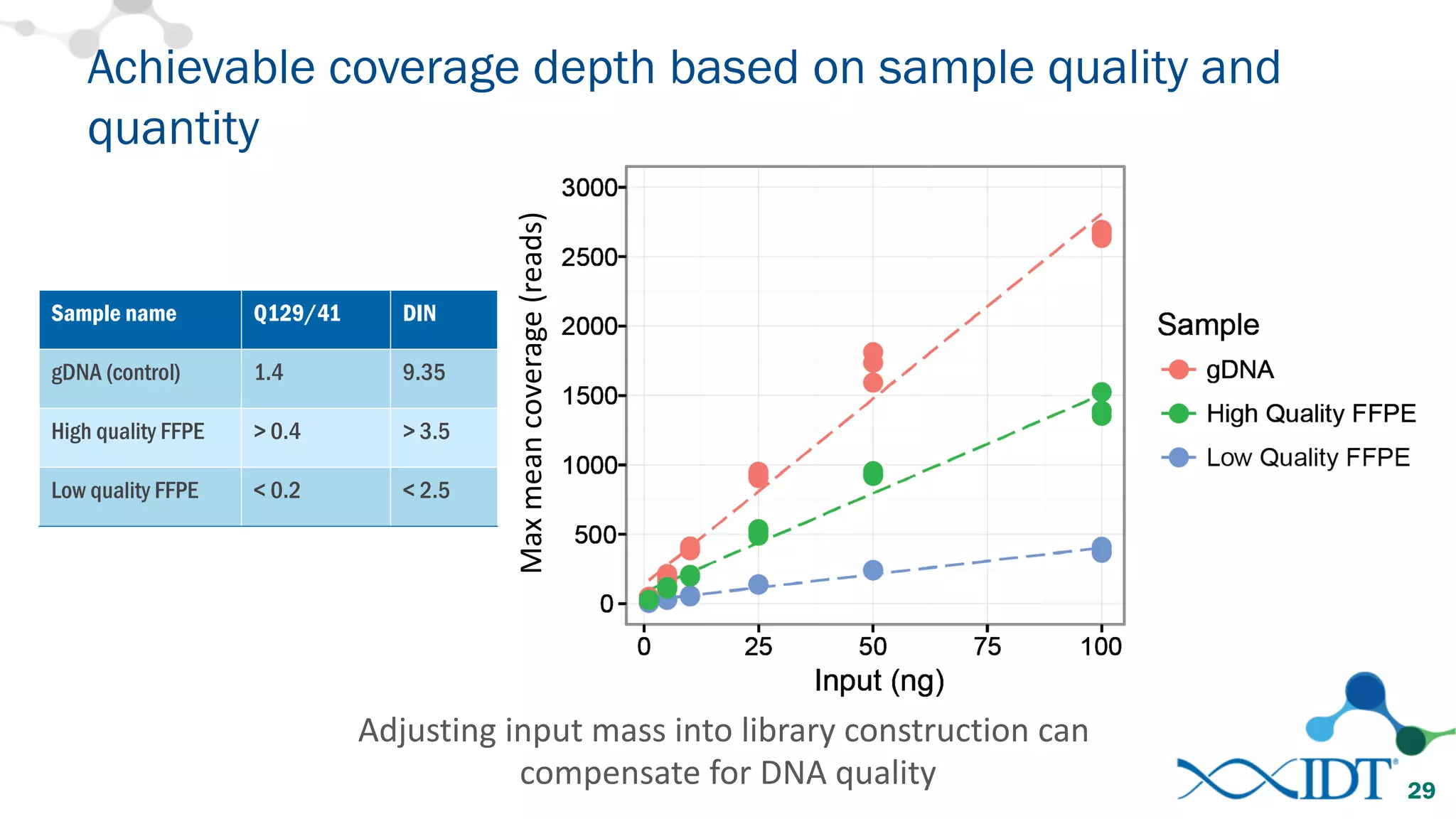
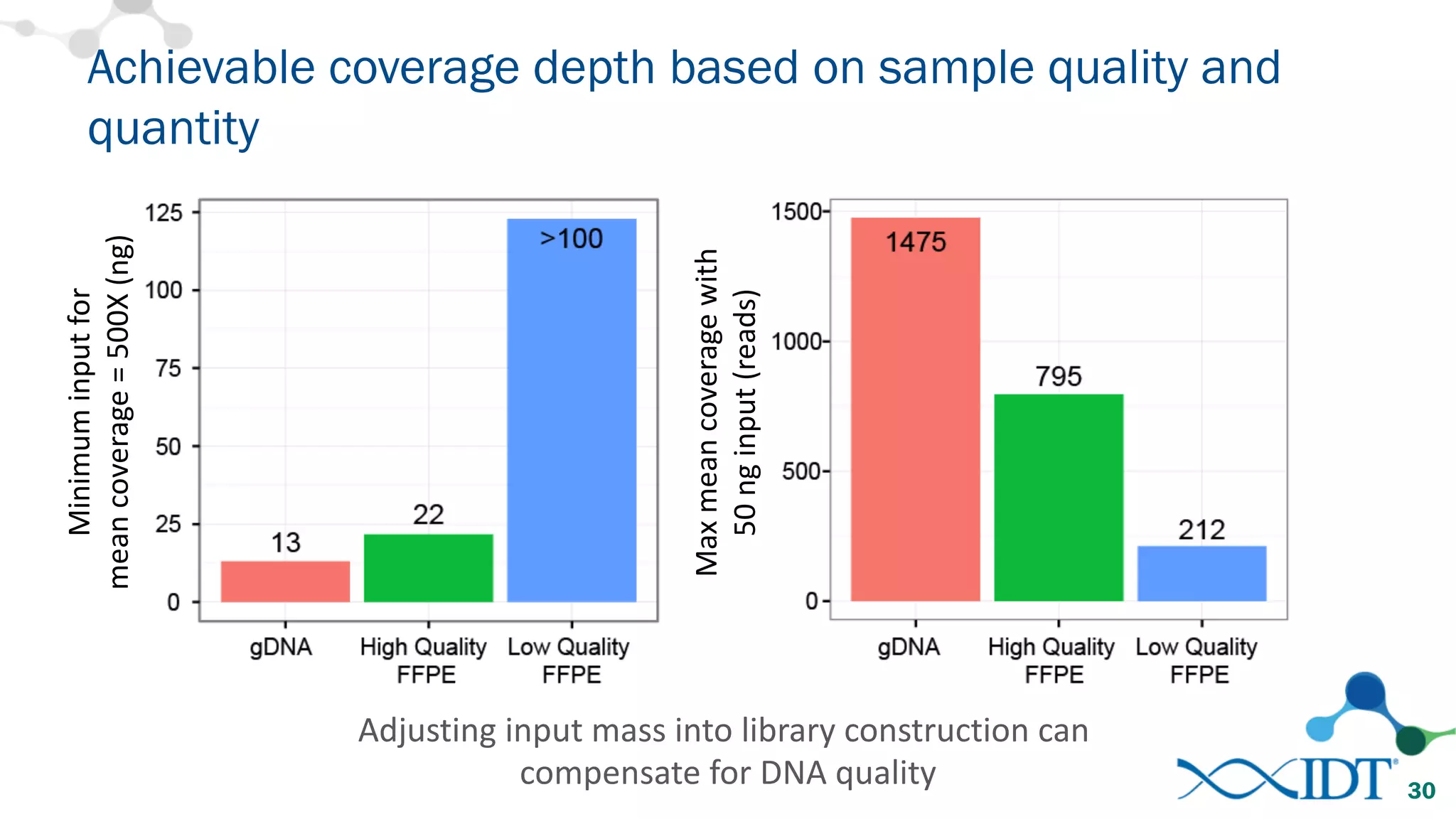
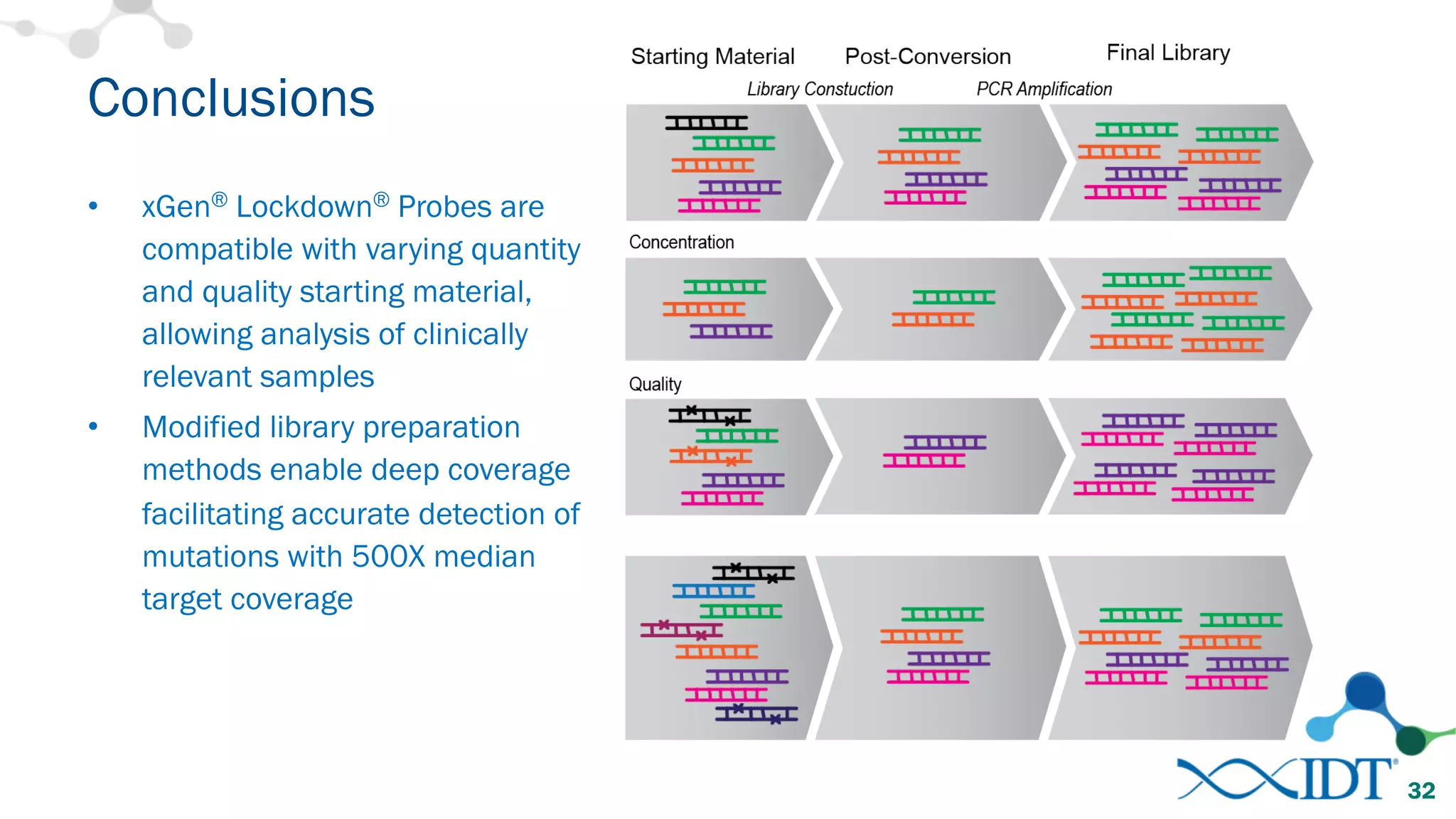
The document provides recommendations for optimizing the capture and sequencing of DNA from formalin-fixed, paraffin-embedded (FFPE) samples, emphasizing the importance of sample quality evaluation and extraction methods. It outlines an experimental approach with phases that assess the variability of extraction kits, the predictiveness of quality control methods, and the feasibility of constructing high-quality capture libraries from FFPE samples. Additionally, it discusses the impact of sample quality and input amounts on library preparation and sequencing performance.