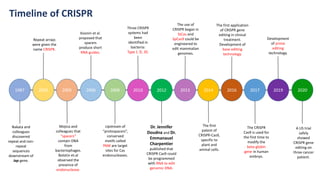
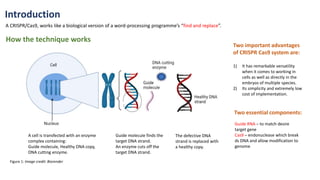
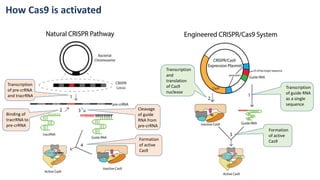
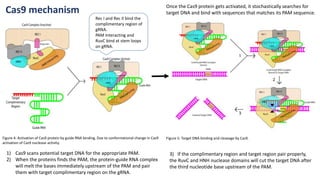
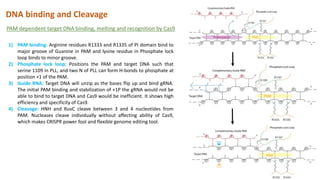
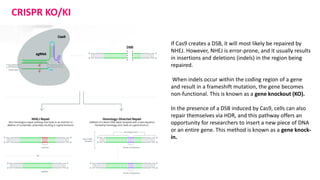
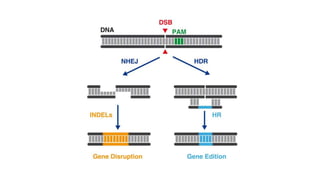
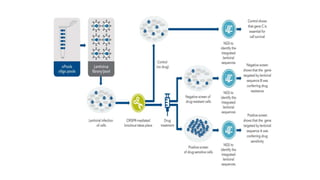
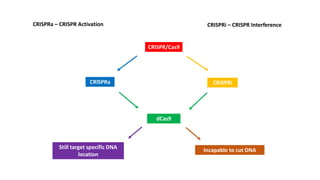
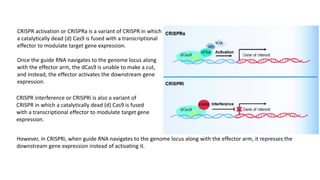
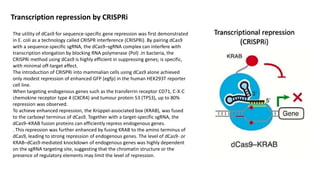
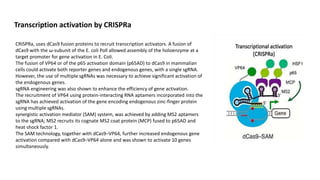
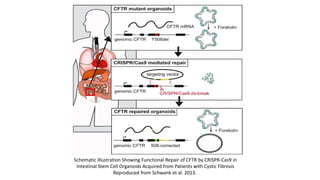
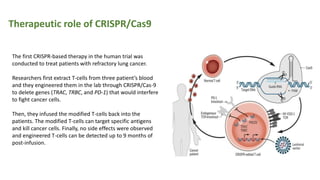
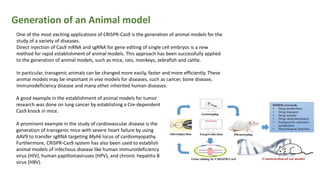
The document elaborates on the discovery and development of CRISPR-Cas9 gene editing technology, including its mechanisms, variants, and applications in various fields. It discusses how CRISPR can be effectively utilized for gene therapy, drug discovery, agriculture, and the creation of animal models to study diseases. Recent advancements like base editing and prime editing, alongside the advantages and versatility of CRISPR, are highlighted, illustrating its potential to treat genetic mutations and enhance biotechnological applications.