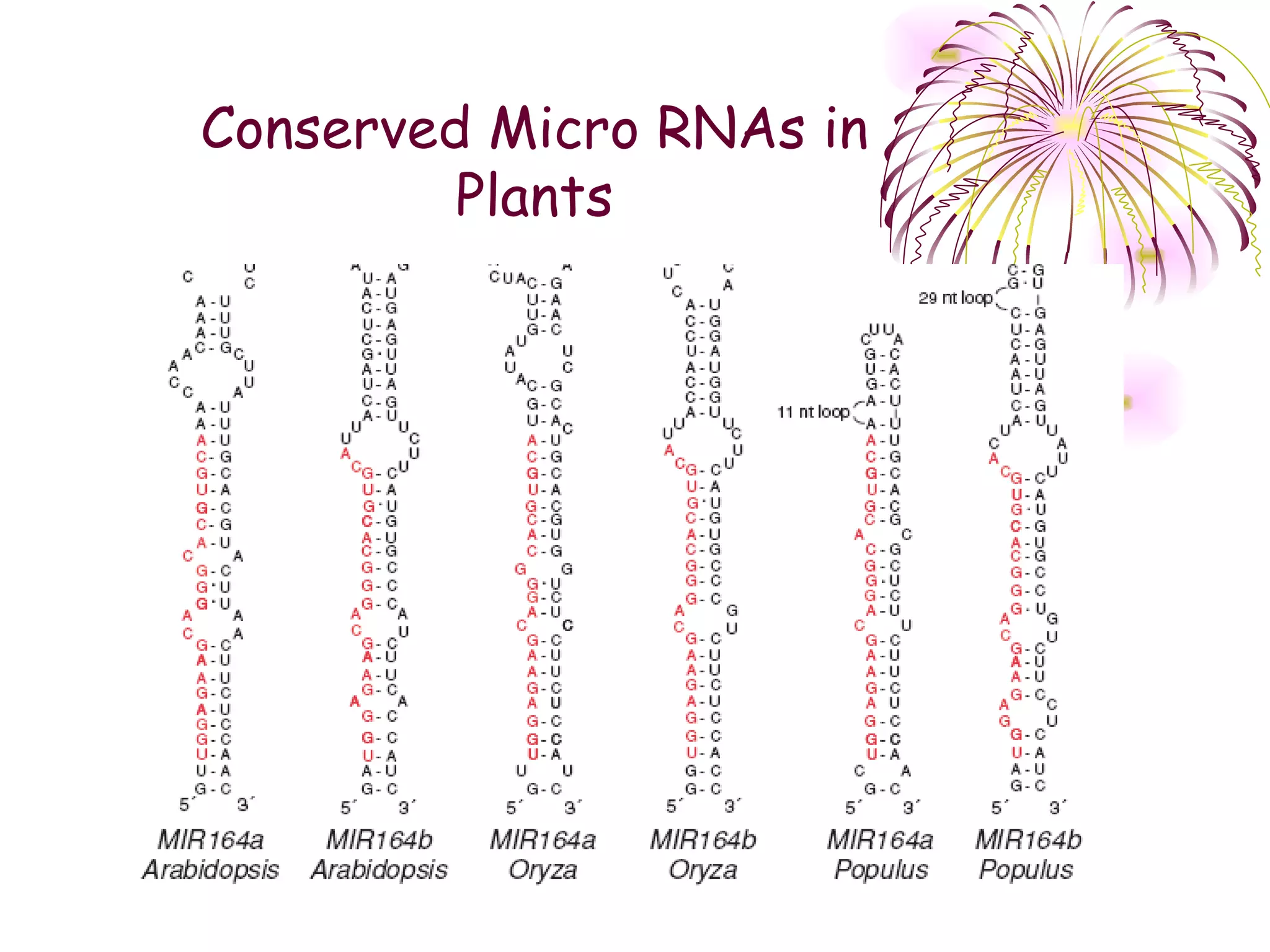
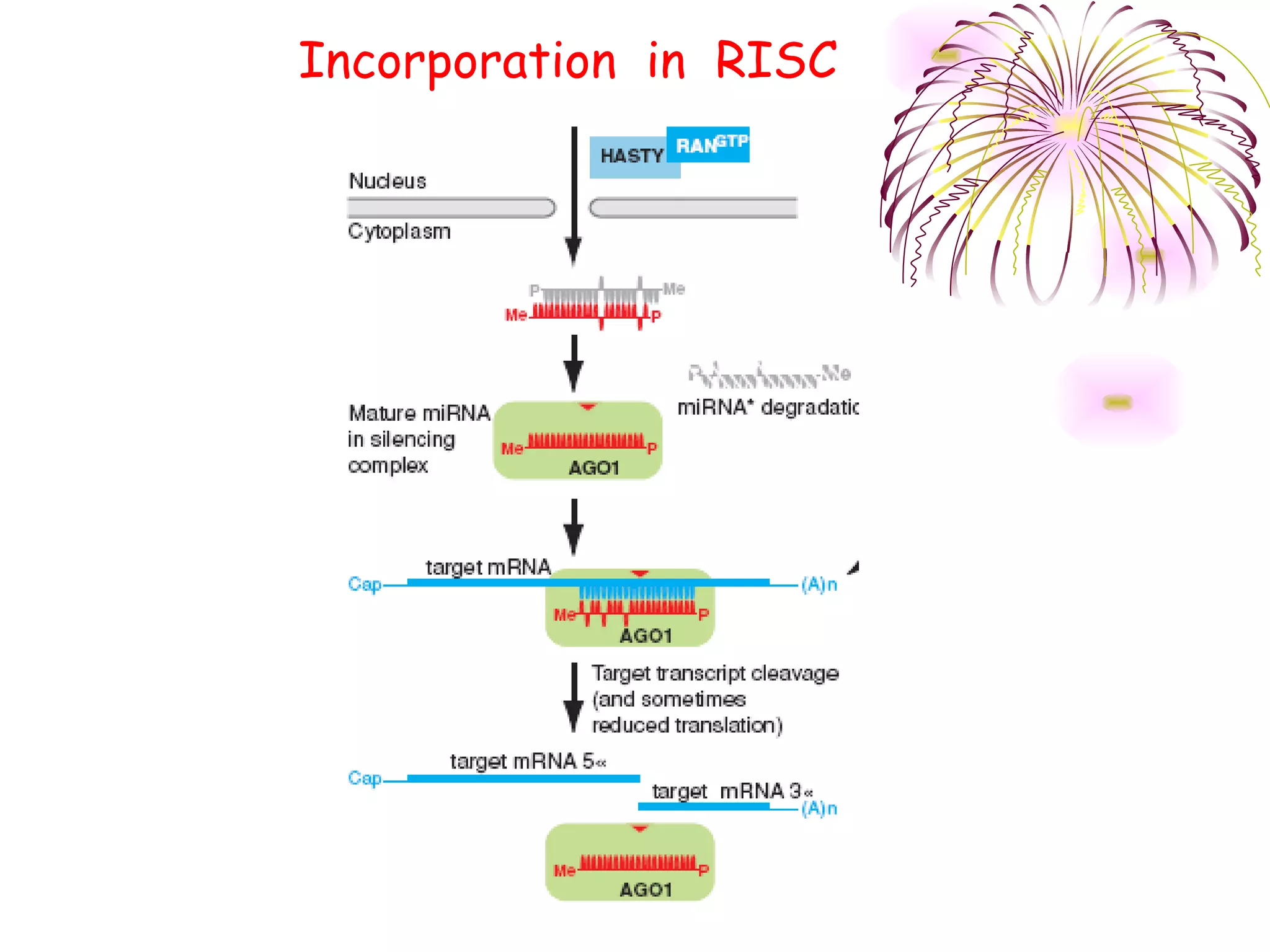
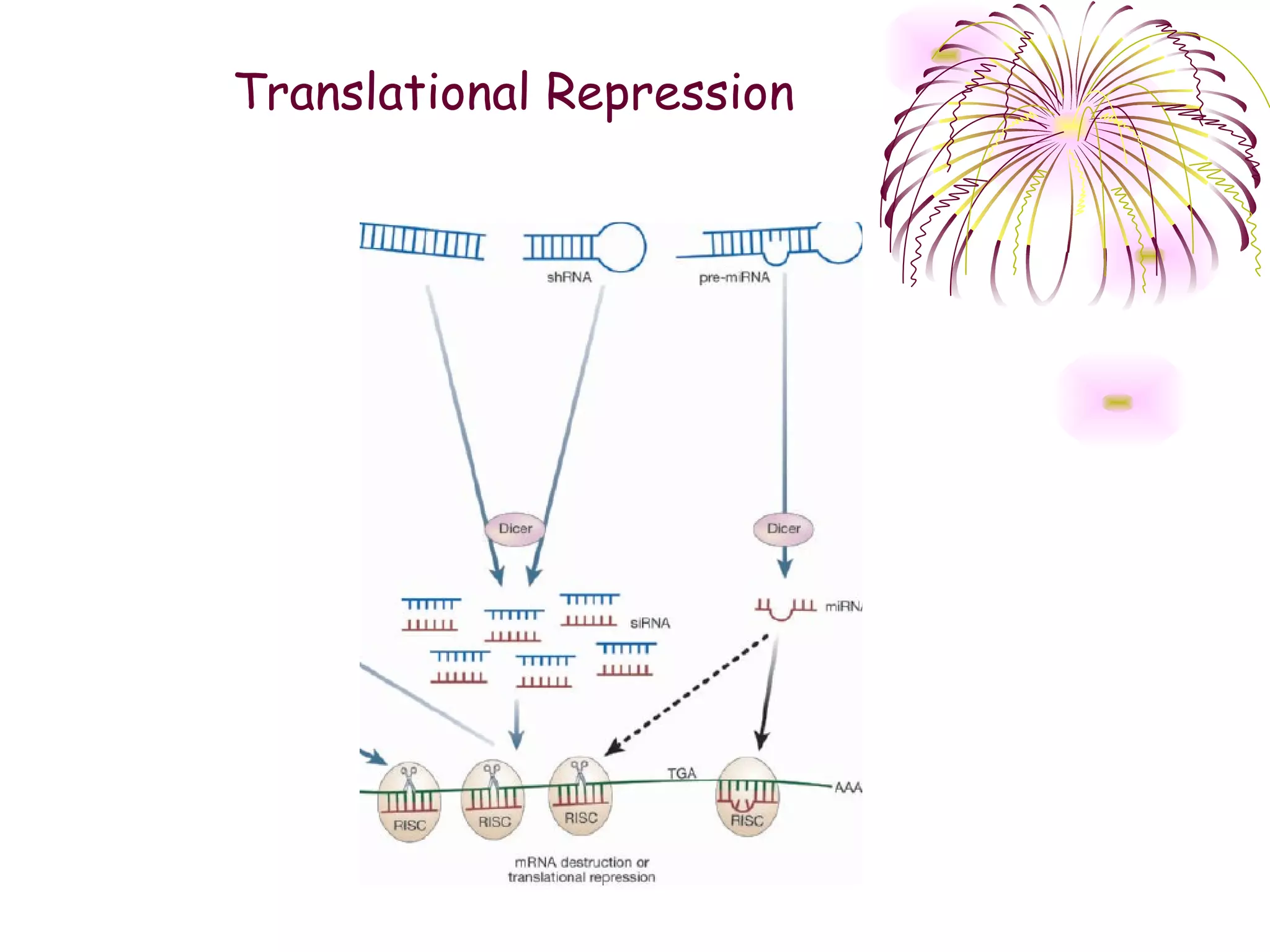
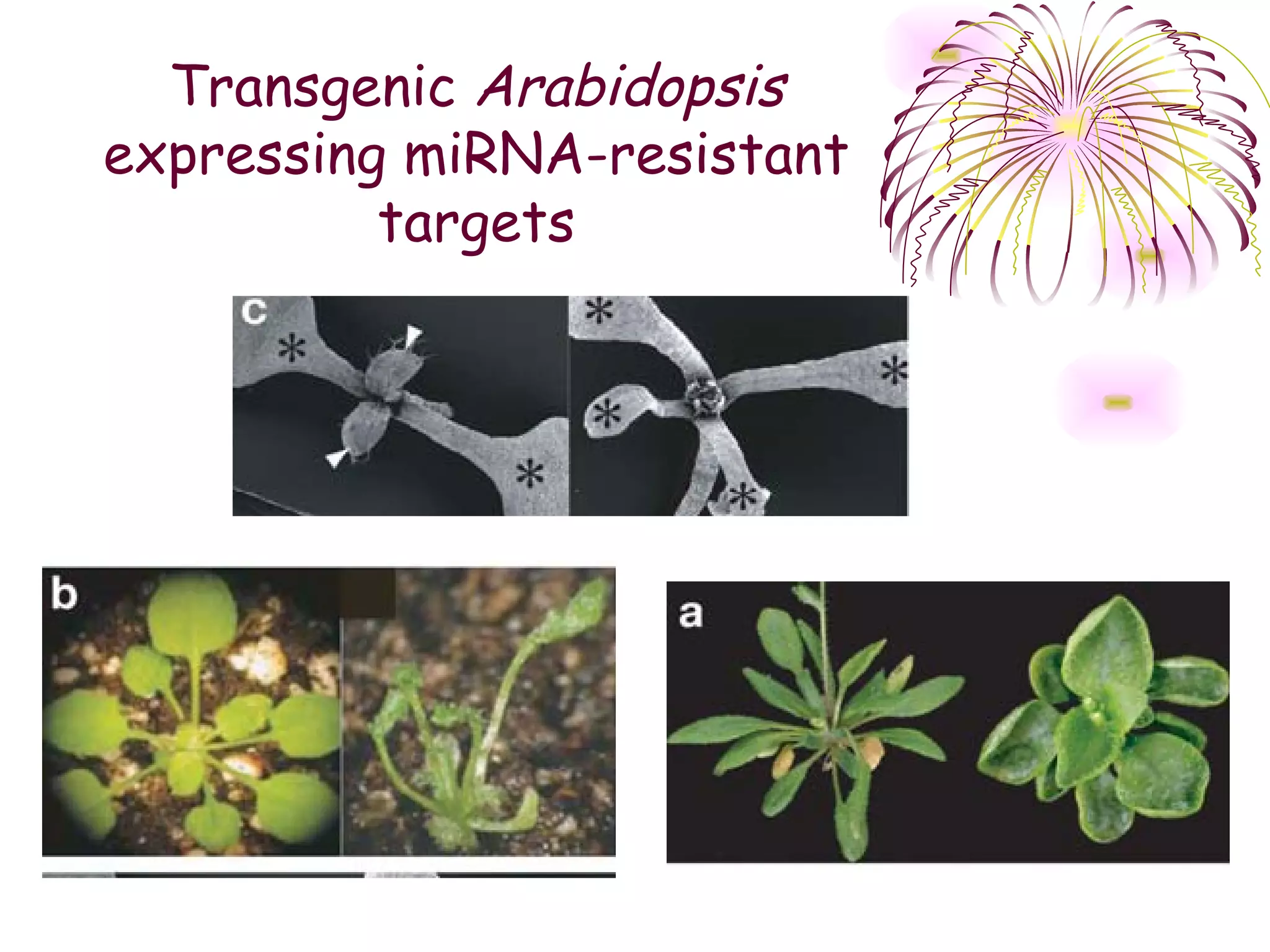
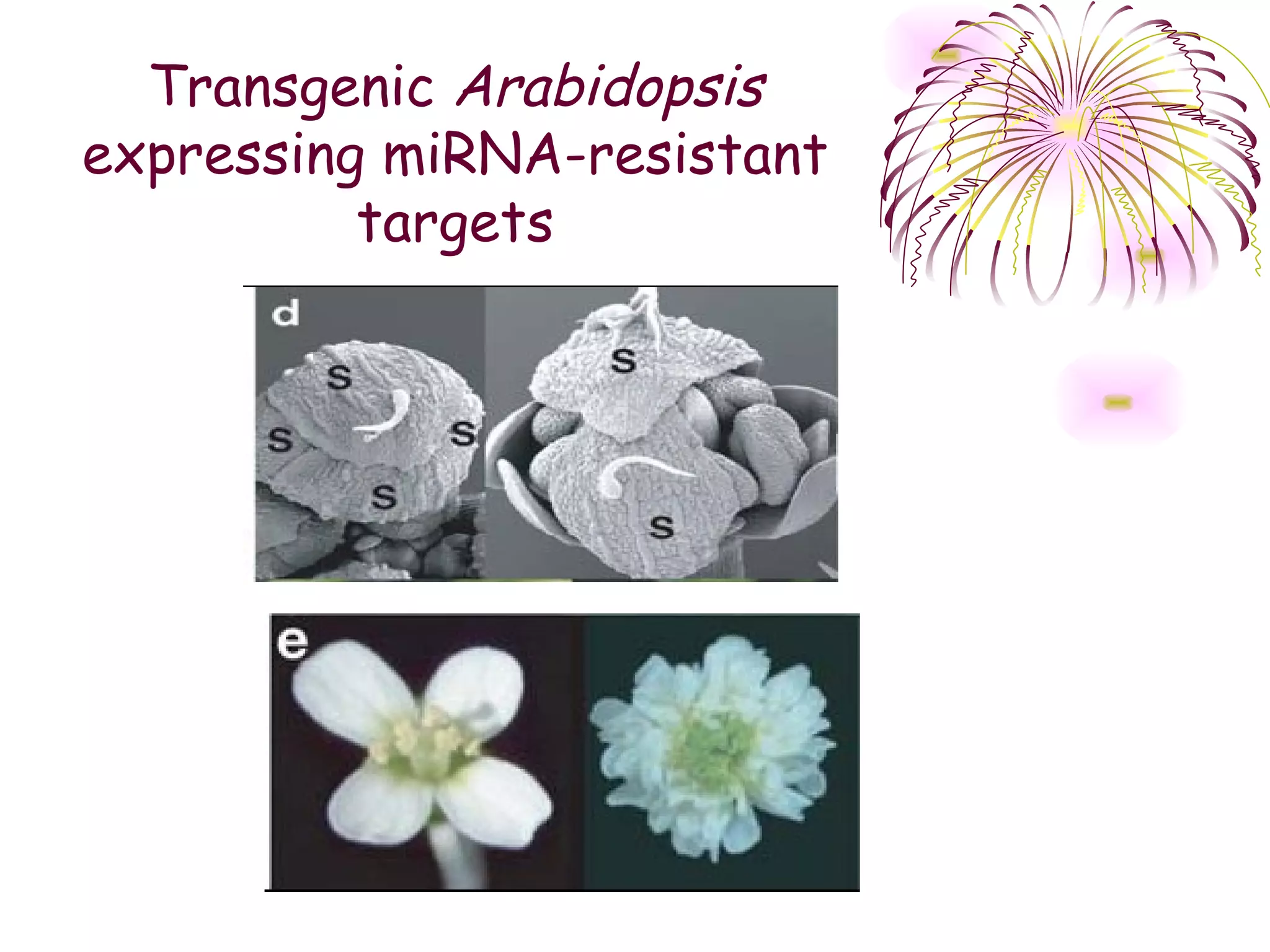
This document discusses microRNAs (miRNAs), which are 22 nucleotide non-coding RNAs that play important regulatory roles in plants. miRNAs are processed from stem-loop precursors by the enzyme Dicer and mediate post-transcriptional gene silencing by guiding mRNA cleavage or translational repression. Bioinformatic and genetic studies have identified many conserved miRNA families in plants and shown that miRNAs regulate key transcription factors to control developmental processes.