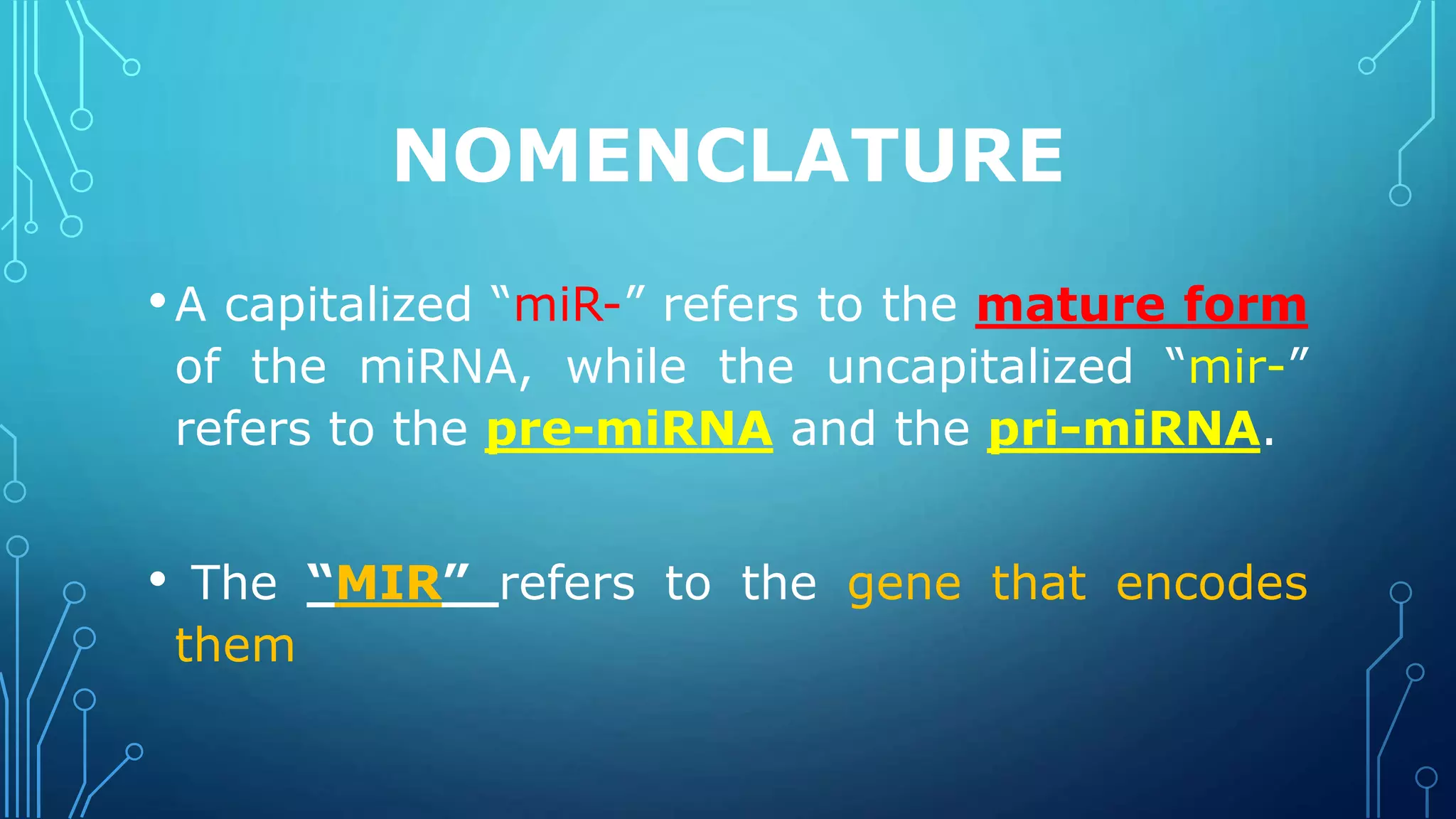
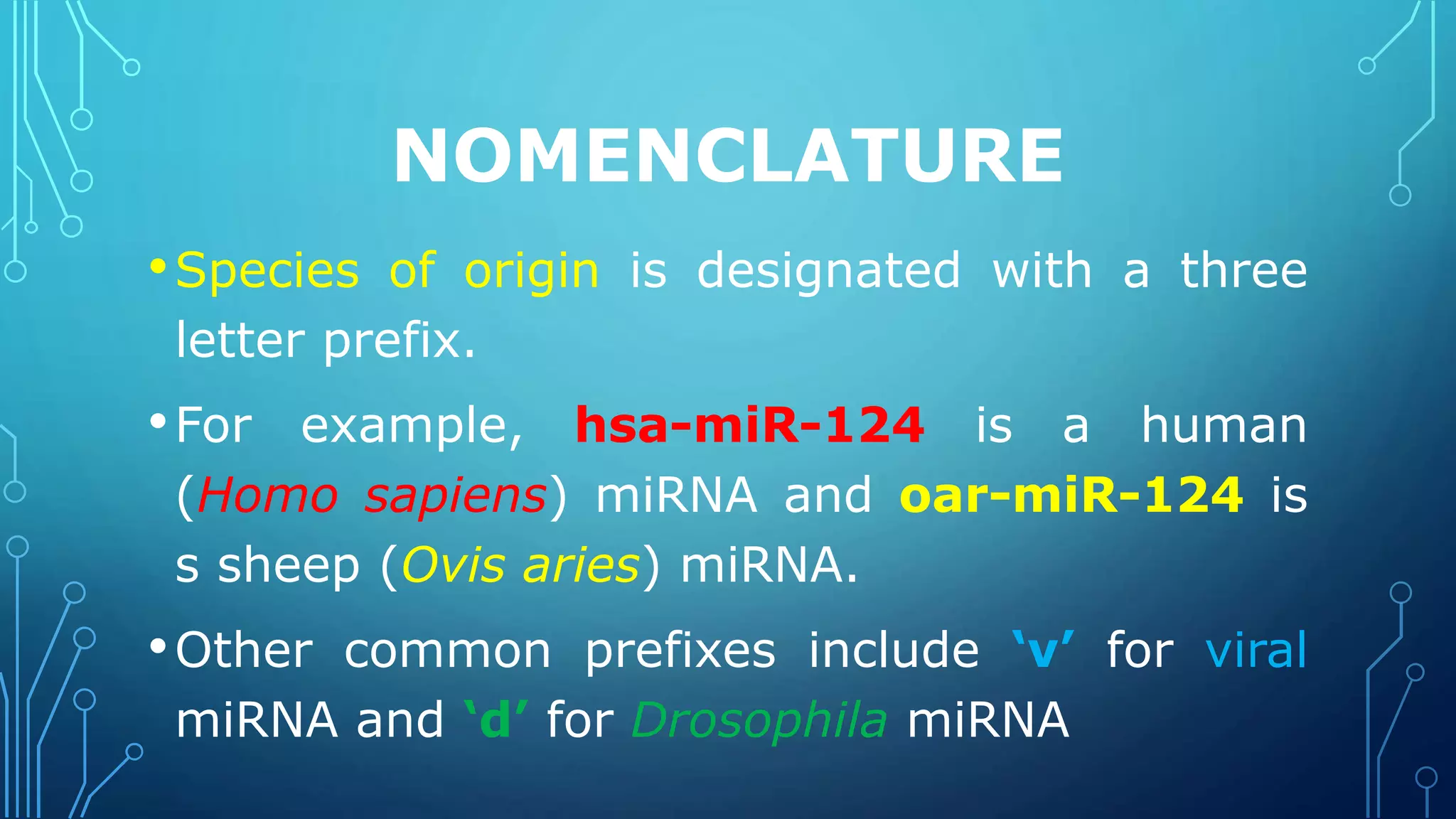
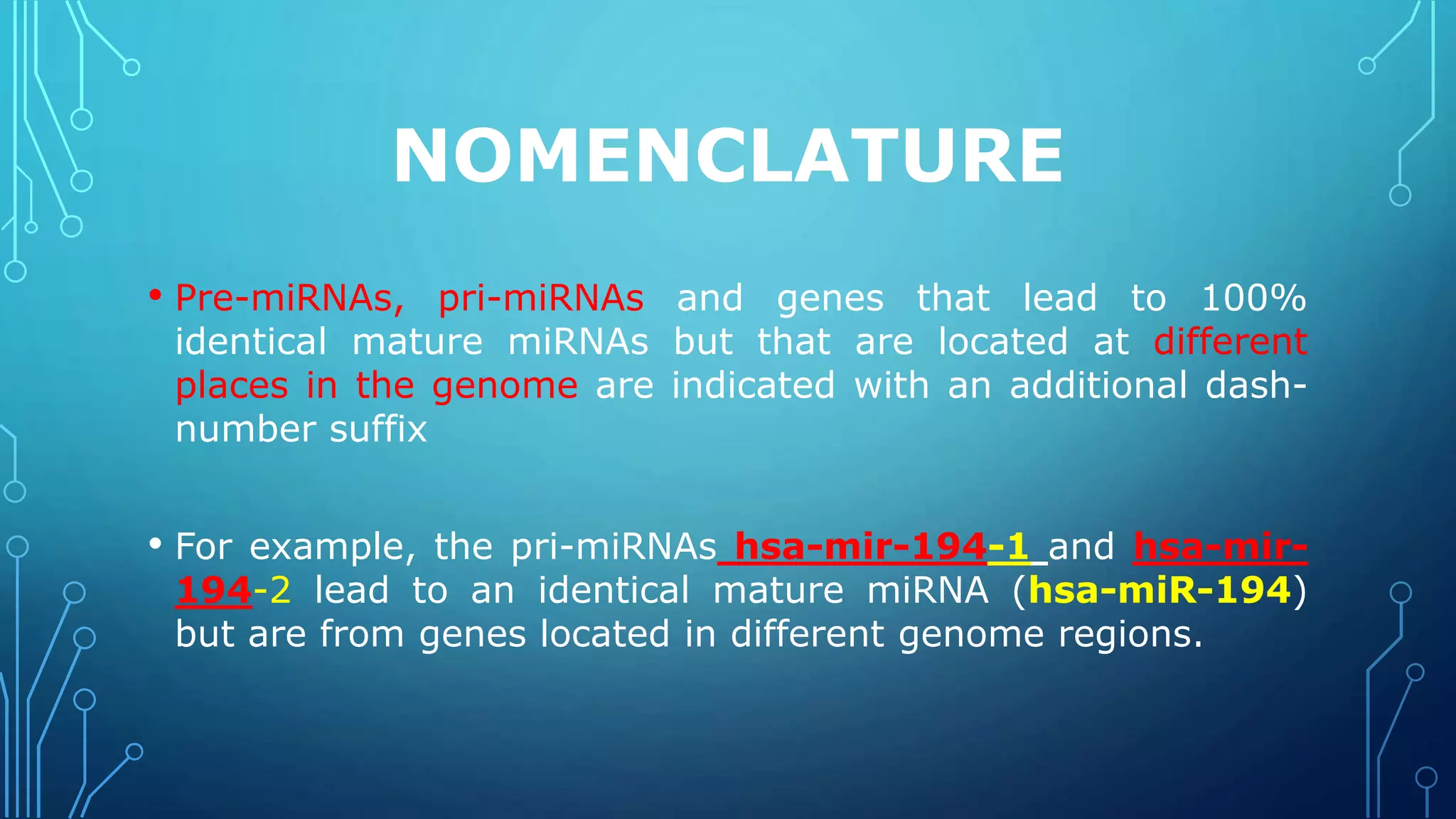
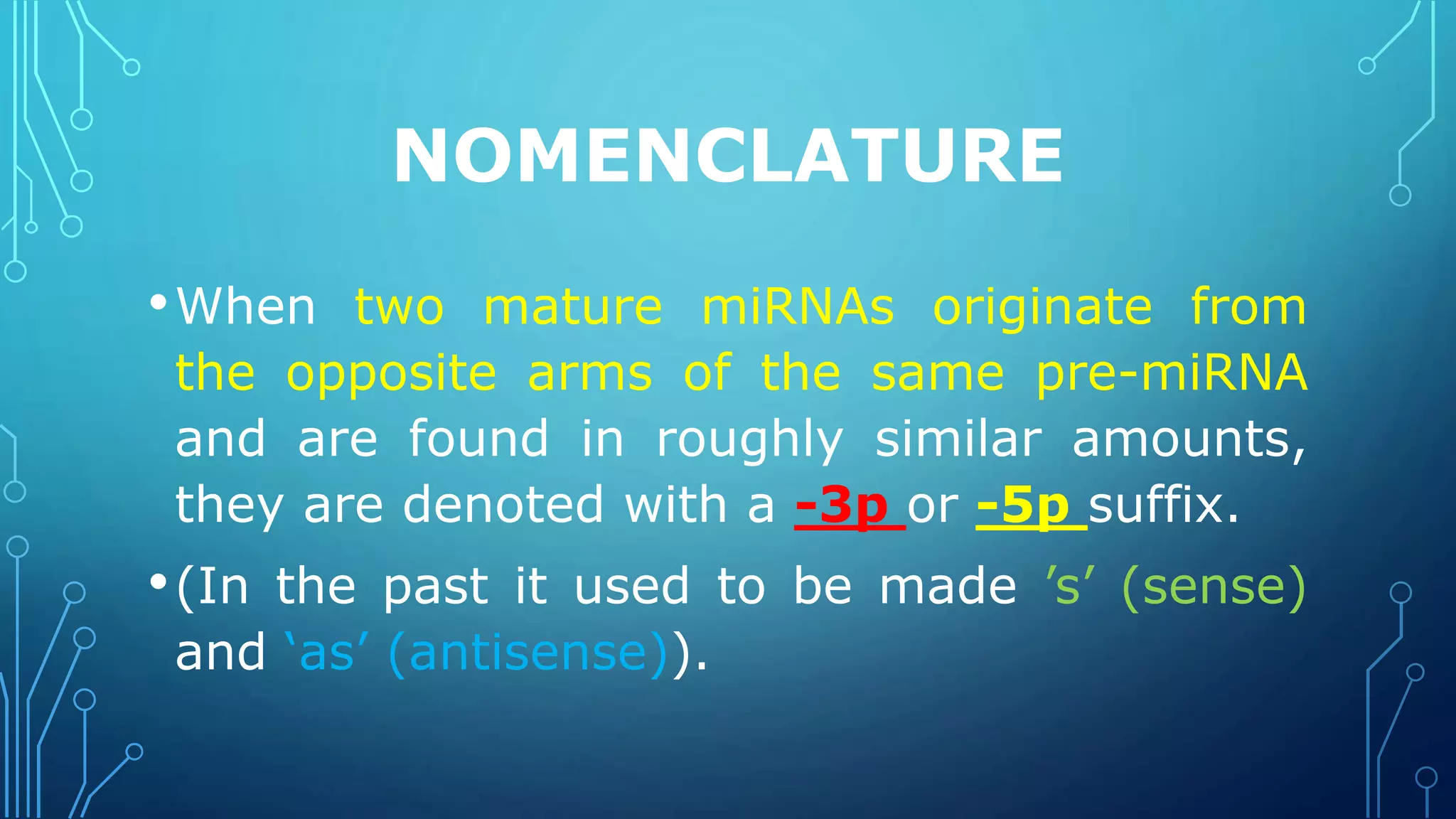
This document discusses microRNAs (miRNAs), including their biogenesis and function. It covers that miRNAs are small noncoding RNAs around 22 nucleotides long that are found in plants, animals and viruses and function in RNA silencing. It then summarizes the key steps in miRNA biogenesis, from transcription of miRNA genes by RNA polymerase II, to nuclear and cytoplasmic processing by the enzymes Drosha, Exportin 5, Dicer and Argonaute to produce the mature miRNA loaded into the RISC complex.