This document discusses several key concepts related to gene structure:
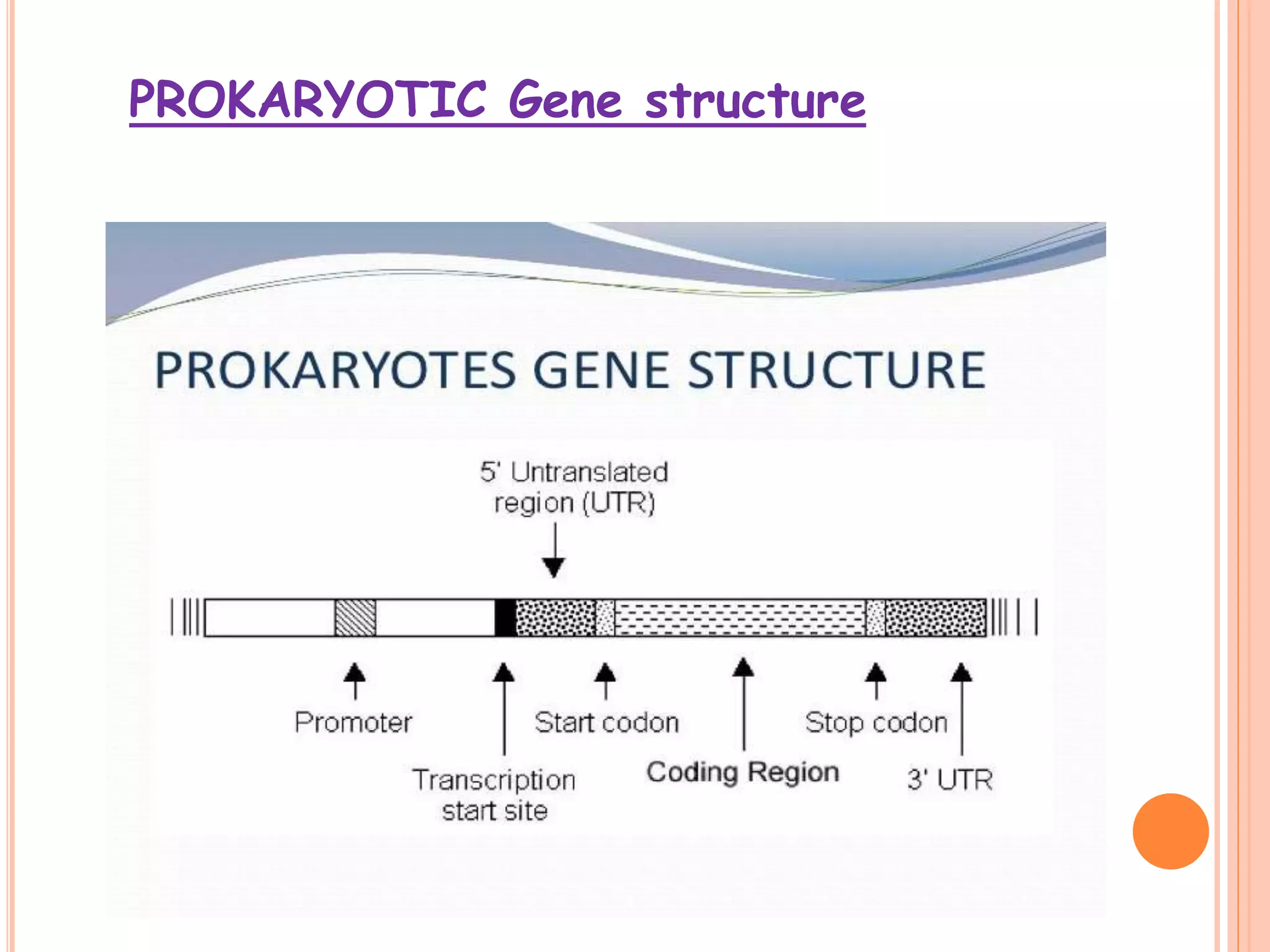
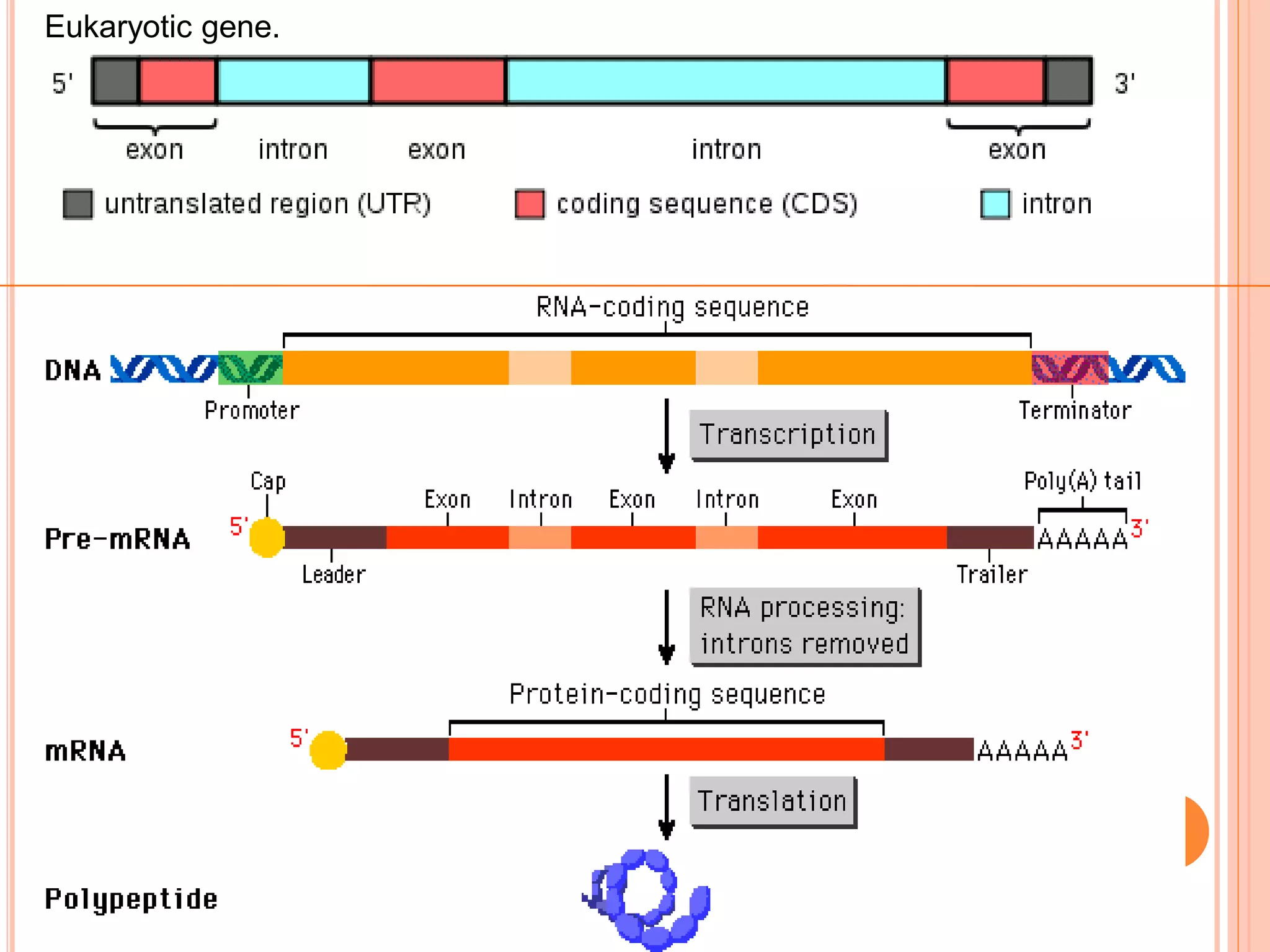
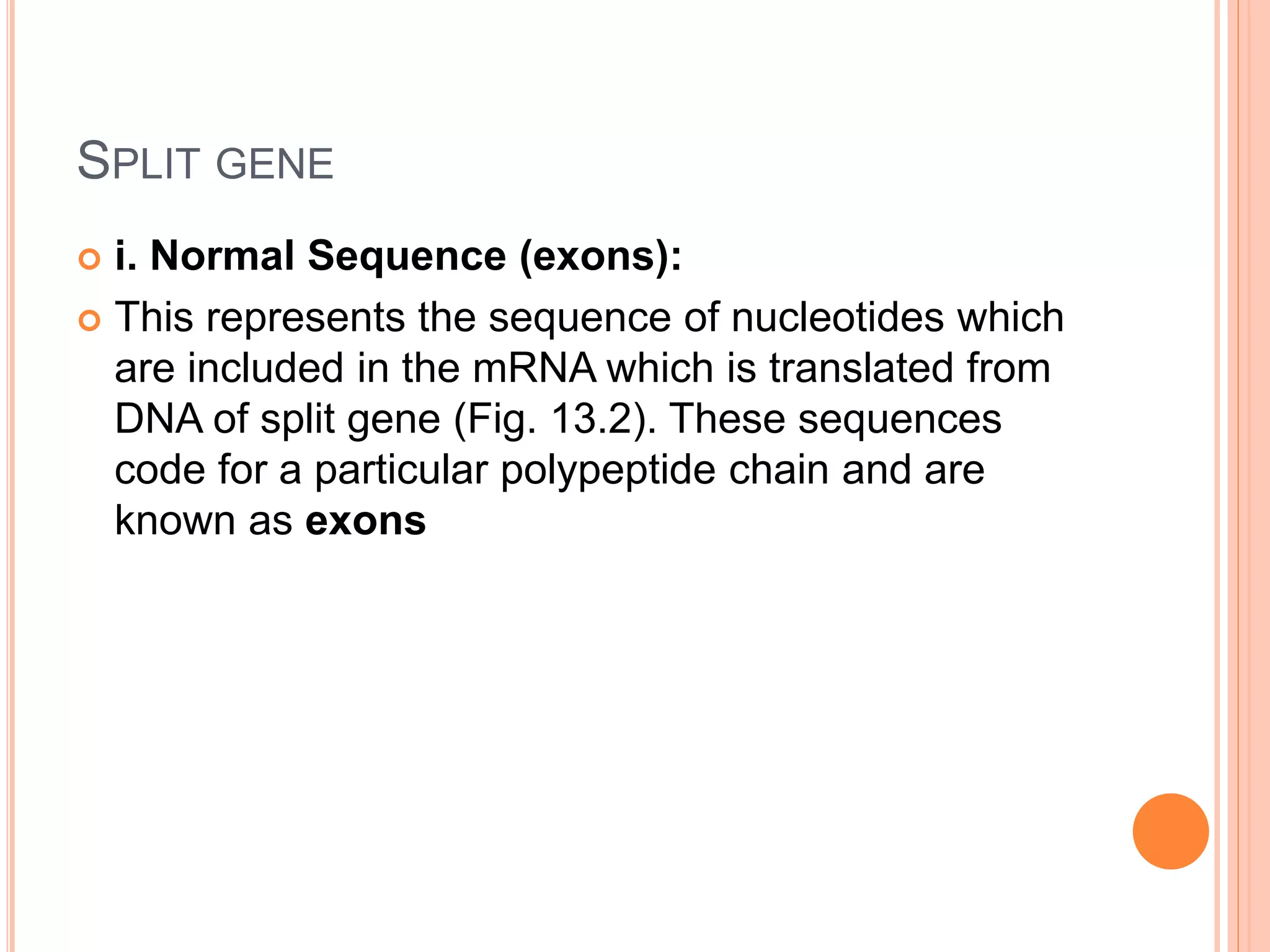
1. Genes in eukaryotes contain both coding (exon) and non-coding (intron) sequences. Introns are removed during RNA processing to form mRNA.
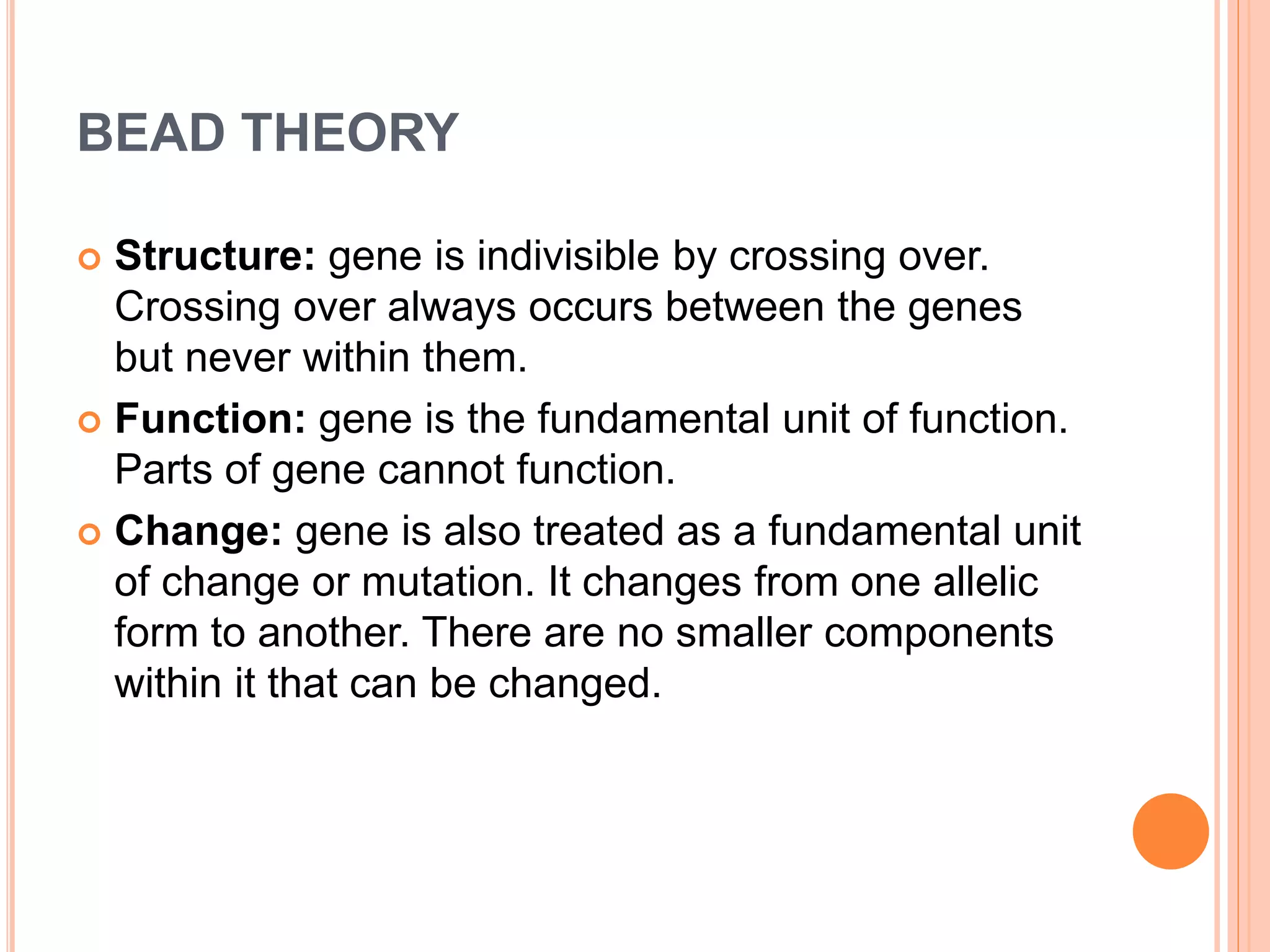
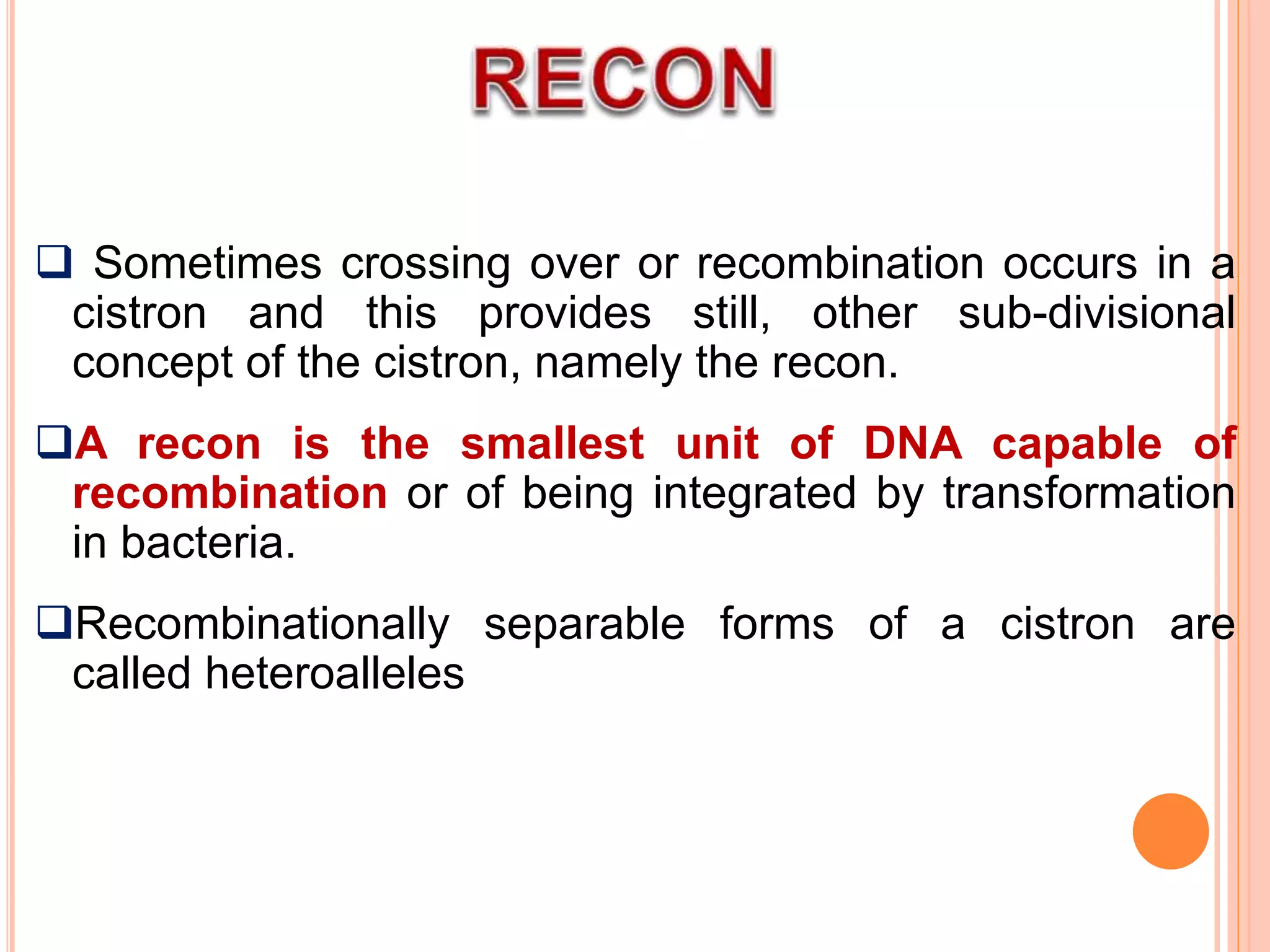
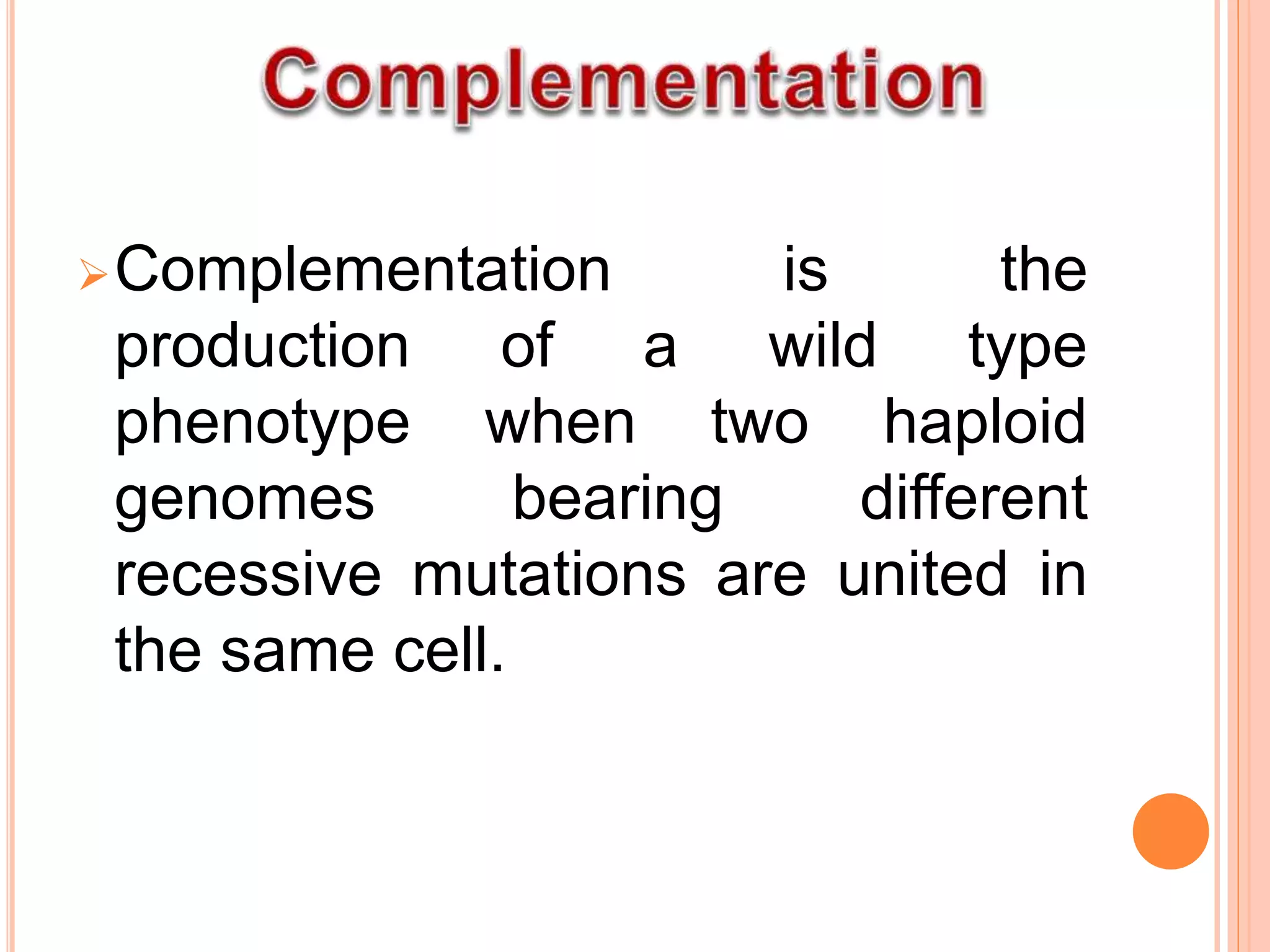
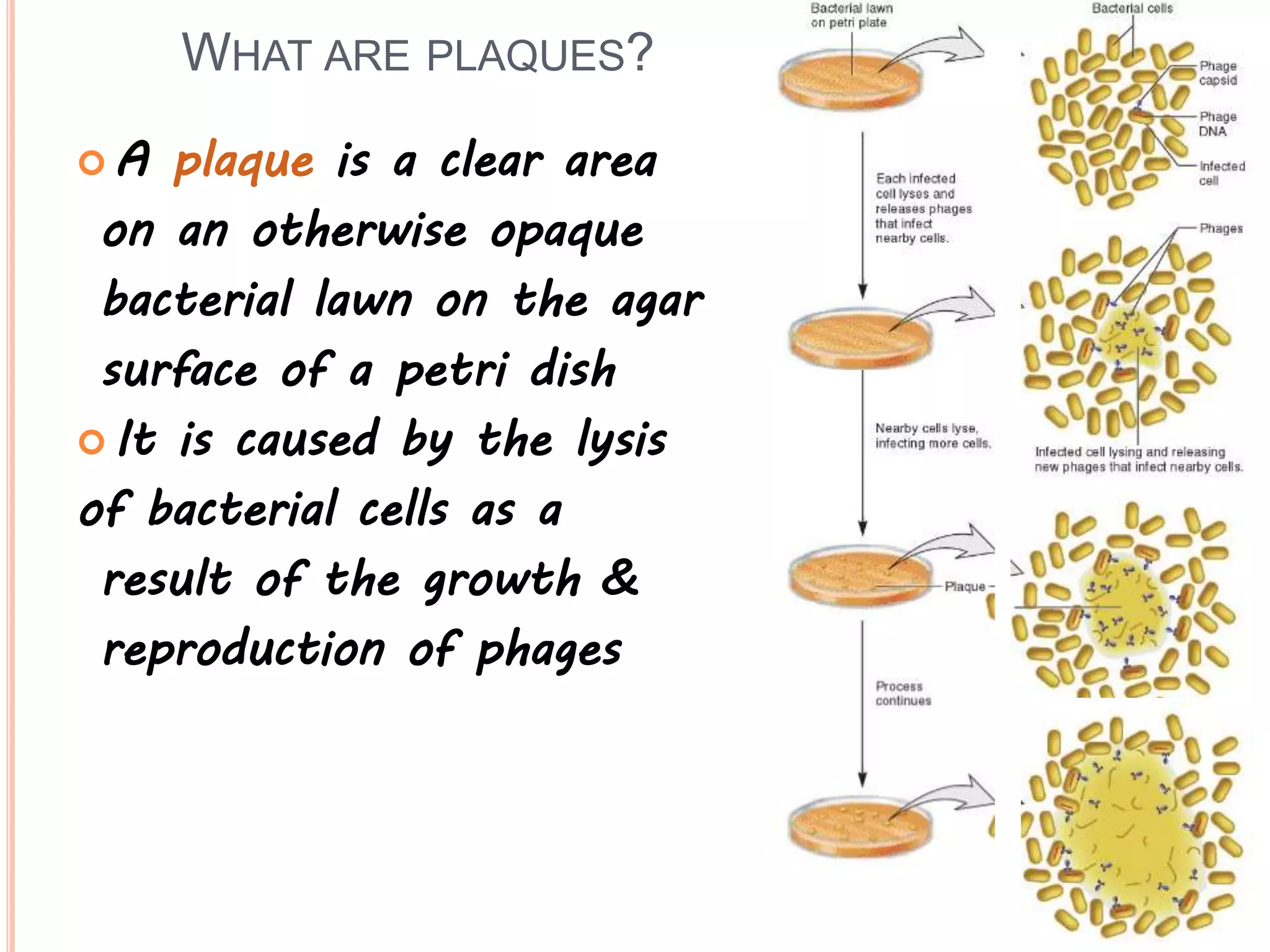
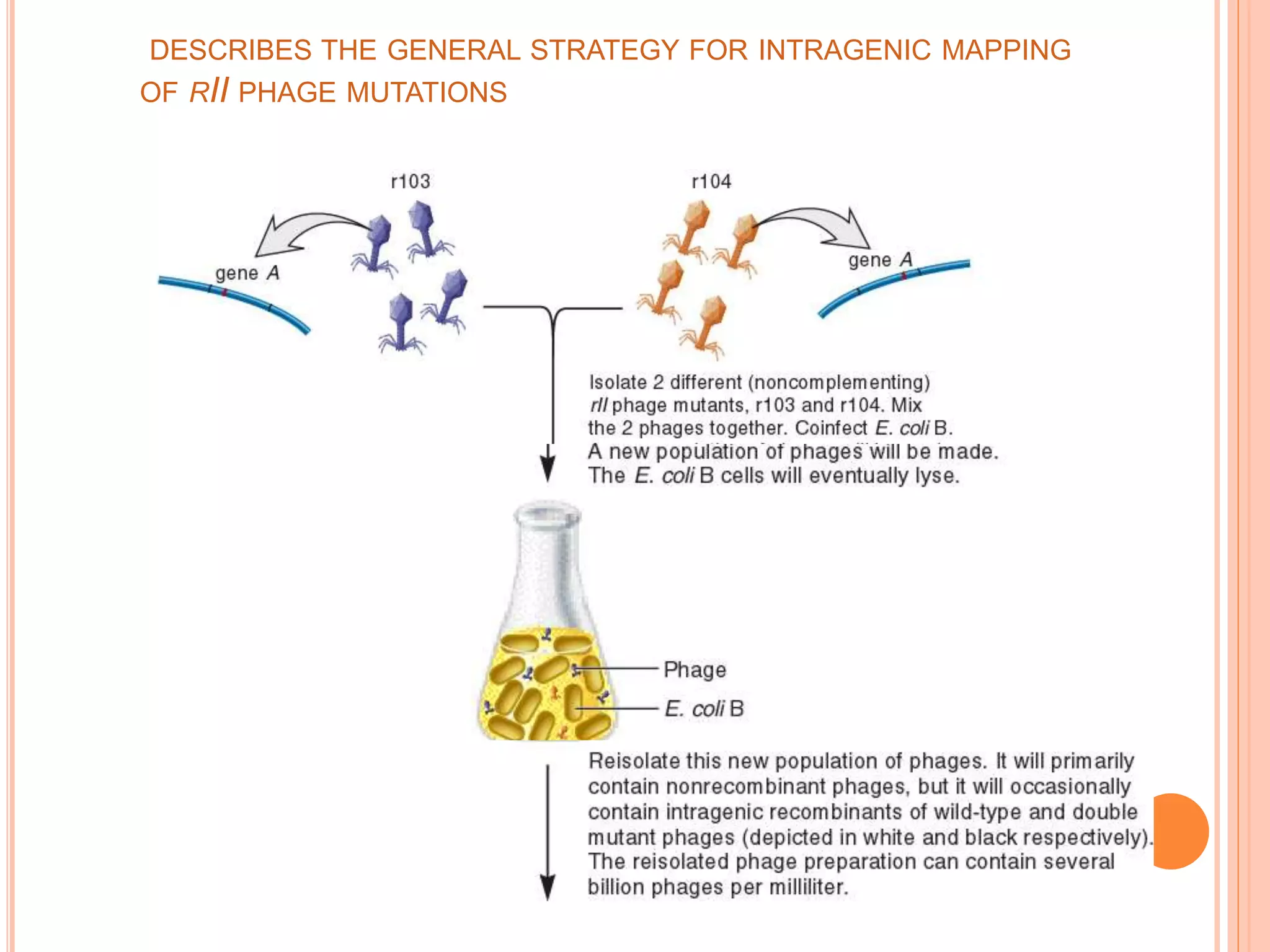
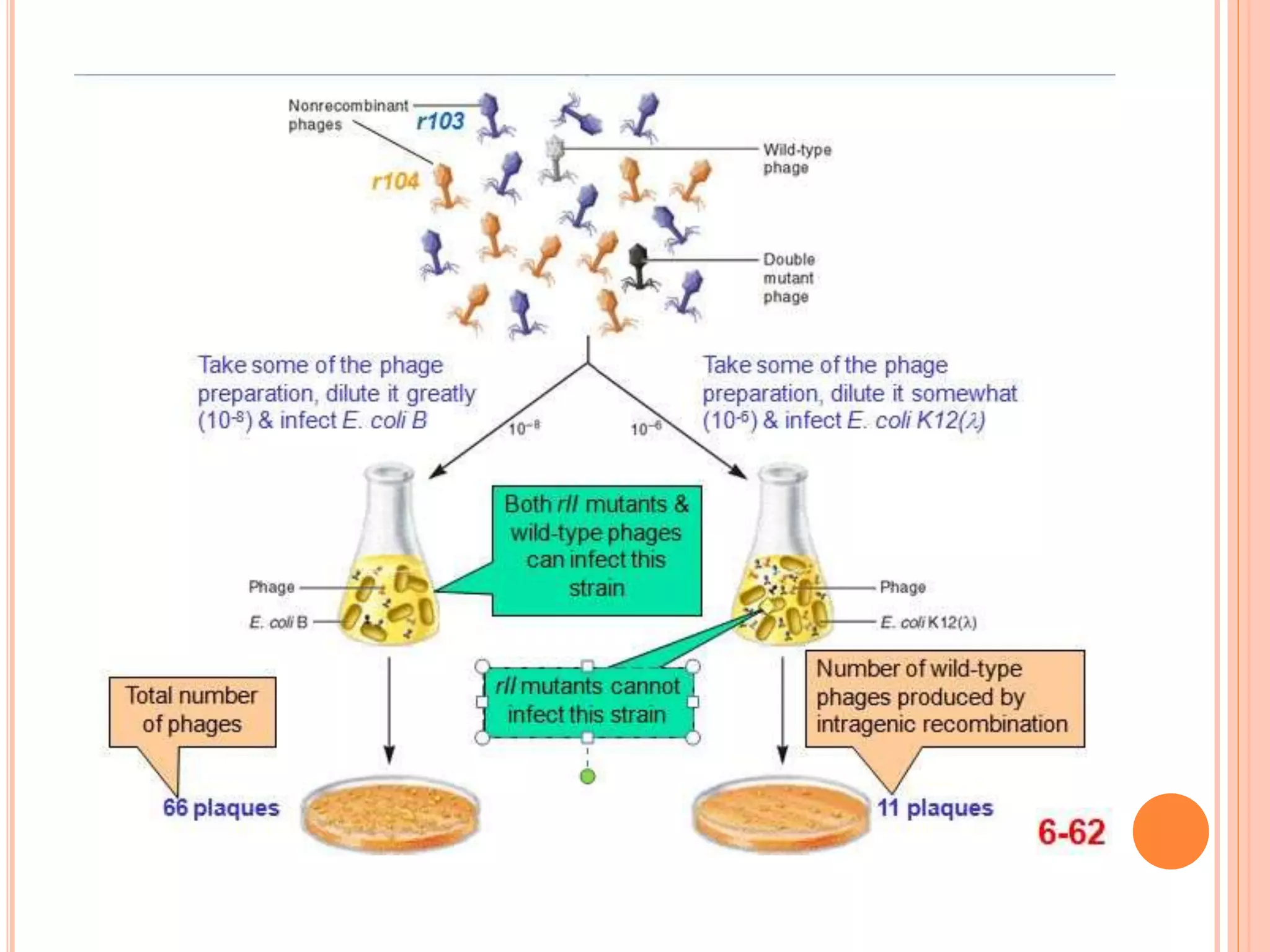
2. Benzer performed fine structure analysis of phage T4 genes which demonstrated that genes can undergo intragenic recombination, or crossing over within the gene. This established that genes have an internal structure smaller than previously believed.
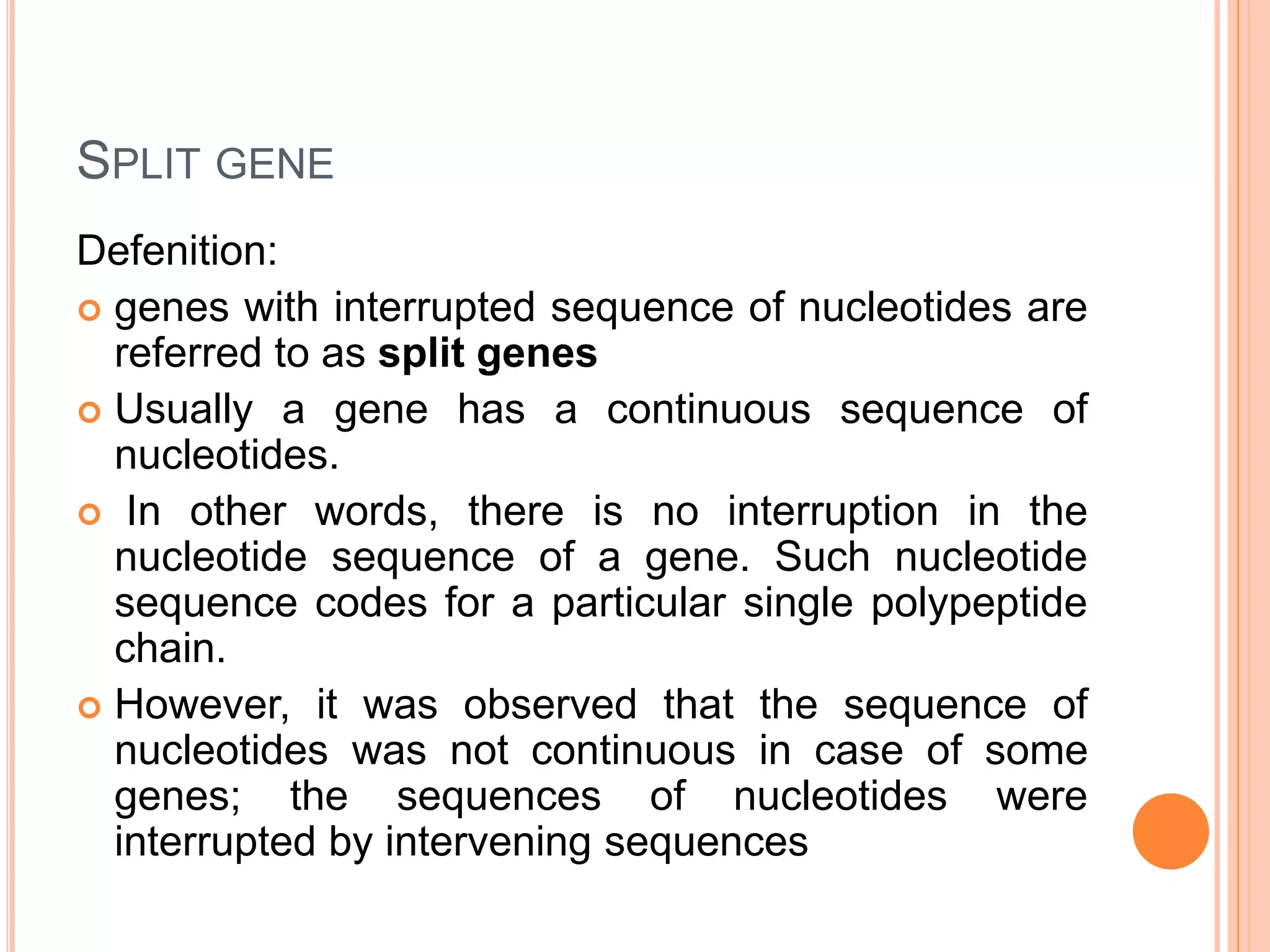
3. Split genes were discovered, which have interrupted sequences (introns) between coding sequences (exons). RNA splicing removes introns to form mature mRNA from split genes.