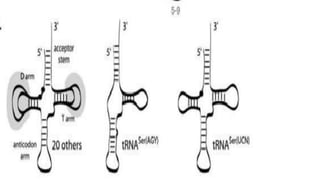
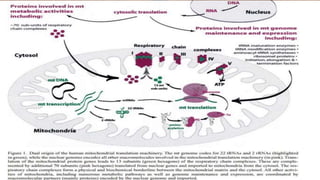
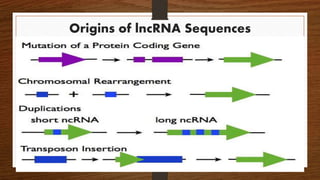
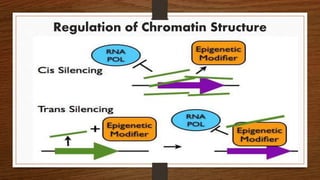
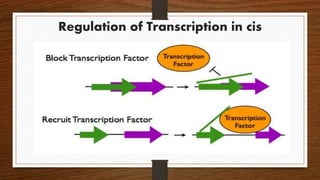
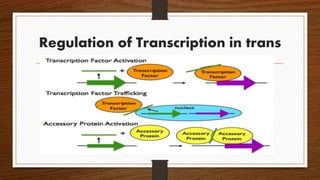
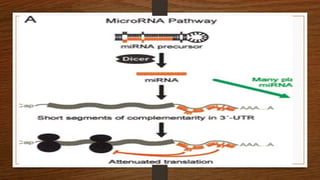
Non-coding RNAs (ncRNAs) are functional RNA molecules that are not translated into proteins. There are several types of ncRNAs that play important roles in biological processes. tRNAs help translate nucleotides into amino acids during protein synthesis. rRNA and snoRNAs are involved in ribosome and RNA structure/modification. MiRNAs regulate gene expression by binding to mRNA. LncRNAs regulate processes like chromatin structure and transcription. Mt-tRNAs specific to mitochondria are essential for oxidative phosphorylation. Mutations can cause diseases like myopathies.