The document provides an overview of various types of RNA, including coding RNA such as messenger RNA (mRNA) and non-coding RNA like ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), and microRNA (miRNA). It details their structures, functions, and processes like RNA splicing and post-transcriptional modifications, as well as the significance of secondary structures in mRNA stability and translation. Additionally, it highlights the roles of long noncoding RNAs (lncRNAs) and piwi-interacting RNAs (piRNAs) in genome stability and physiological processes.
![Types of RNA:
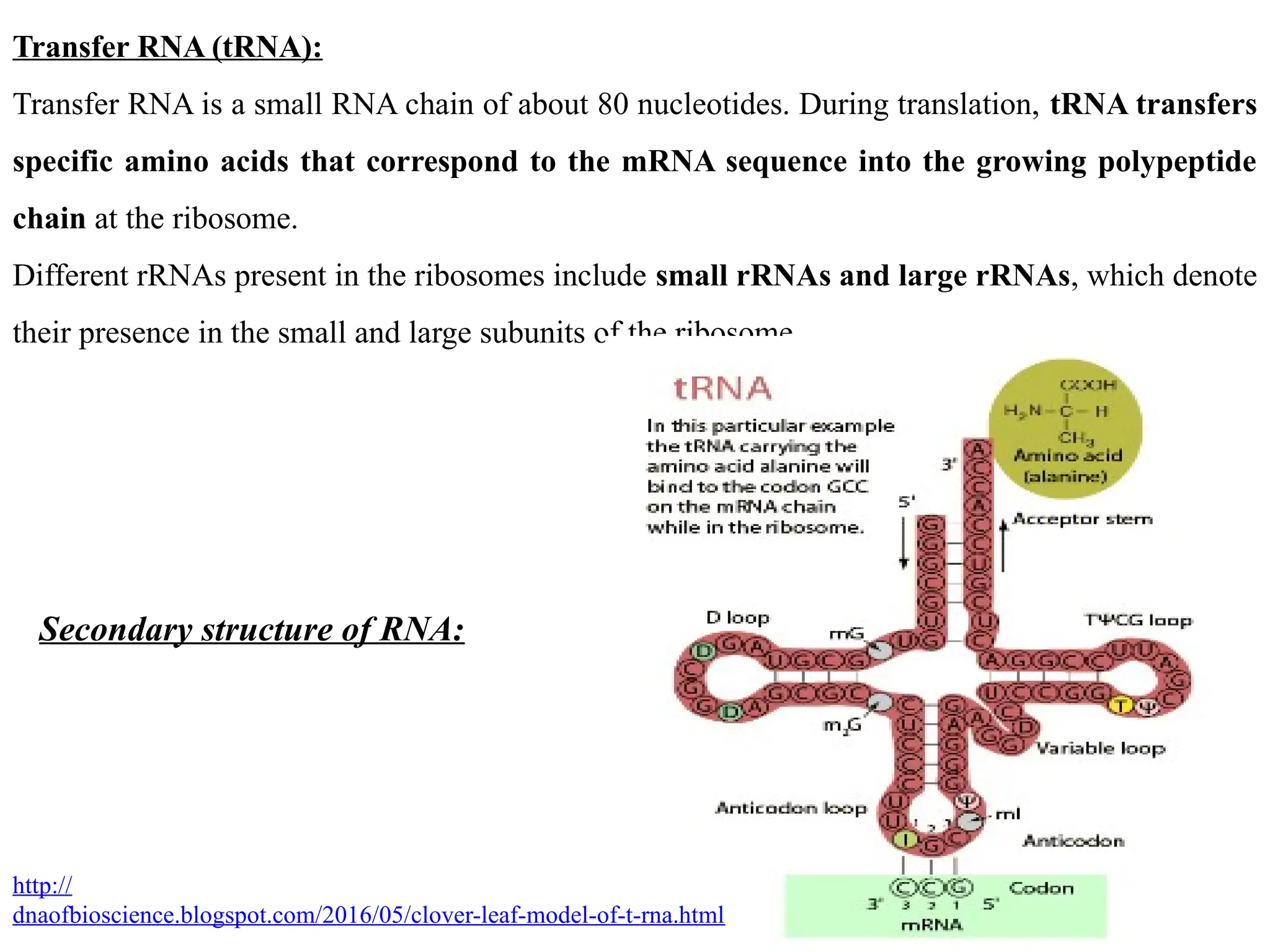
RNA:
RNA or ribonucleic acid is a polymer of nucleotides which is made up of a ribose sugar, a
phosphate, and bases such as adenine, guanine, cytosine, and uracil.
It is a polymeric molecule essential in various biological roles in coding, decoding, regulation,
and expression of genes.
RNA
Coding-RNA Non-coding RNA
messenger RNA[mRNA] Ribosomal RNA (rRNA)
Transfer RNA (tRNA)
Small nuclear RNAs (snRNA; 150 nt)
Small nucleolar RNAs (snoRNA;60-300 nt)
Piwi-interacting RNAs
MicroRNAs (miRNA; 21-22 nt)
Long noncoding RNAs](https://image.slidesharecdn.com/typesofrna-241113092306-9d01579c/75/Types-of-RNA-structure-and-its-functions-pptx-1-2048.jpg)
![Non-coding RNA (ncRNA)
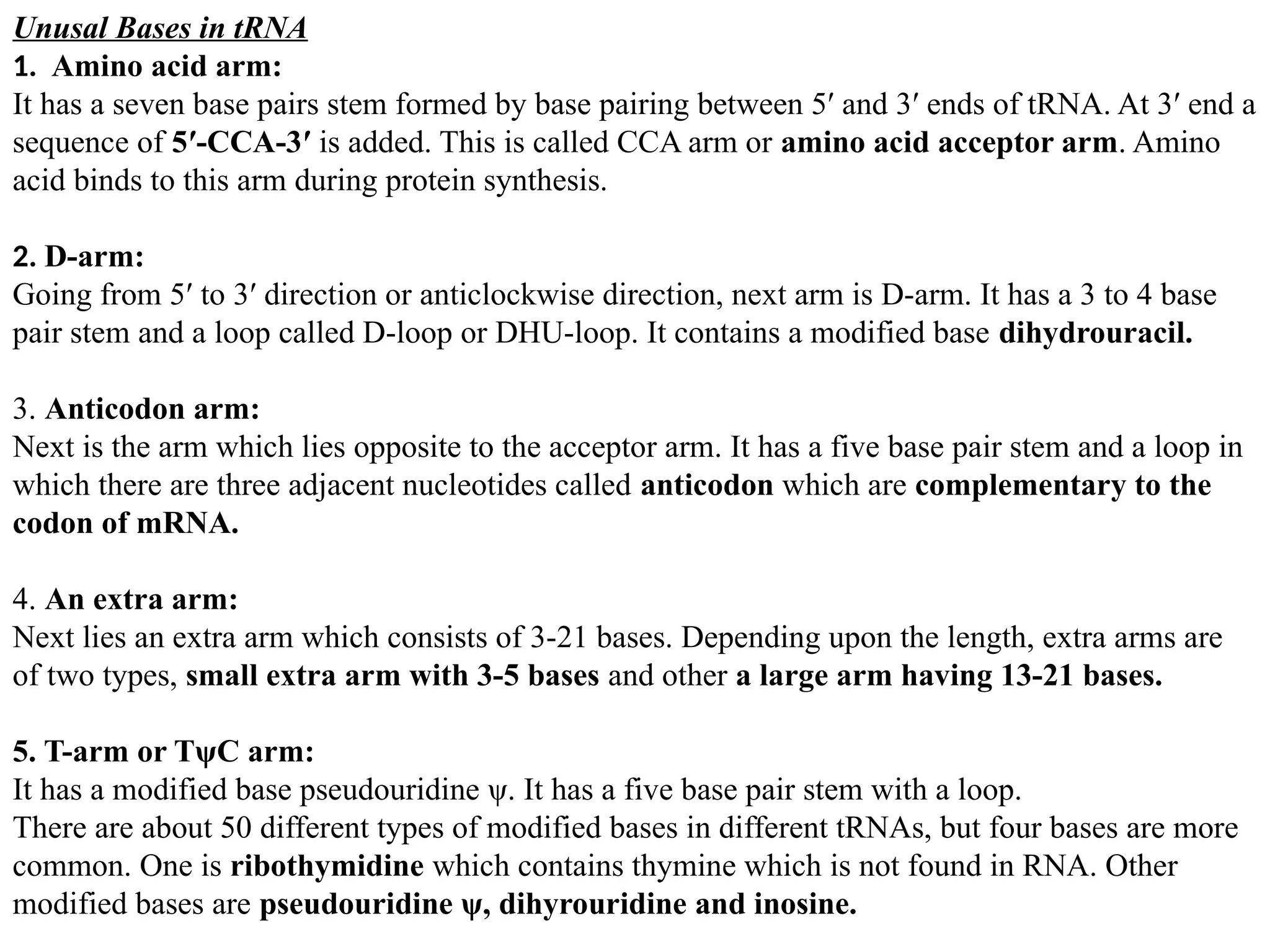
Ribosomal RNA (rRNA):
Ribosomal RNA is the catalytic component of the ribosomes. In the cytoplasm, rRNAs and
protein components combine to form a nucleoprotein complex called the ribosome which binds
mRNA and synthesizes proteins (also called translation).
Table: 1 Composition of Eukaryotic Ribosome
80S [mass = 4220kD]
40S 60S
18S rRNA 28S rRNA
33 Different proteins 5.8S rRNA
5S rRNA
[49 Different proteins]
Courtesy: Biochemistry by Reginald H. Garrett, Charles M. Grisham (4th ed.)
Page No. 962](https://image.slidesharecdn.com/typesofrna-241113092306-9d01579c/75/Types-of-RNA-structure-and-its-functions-pptx-3-2048.jpg)
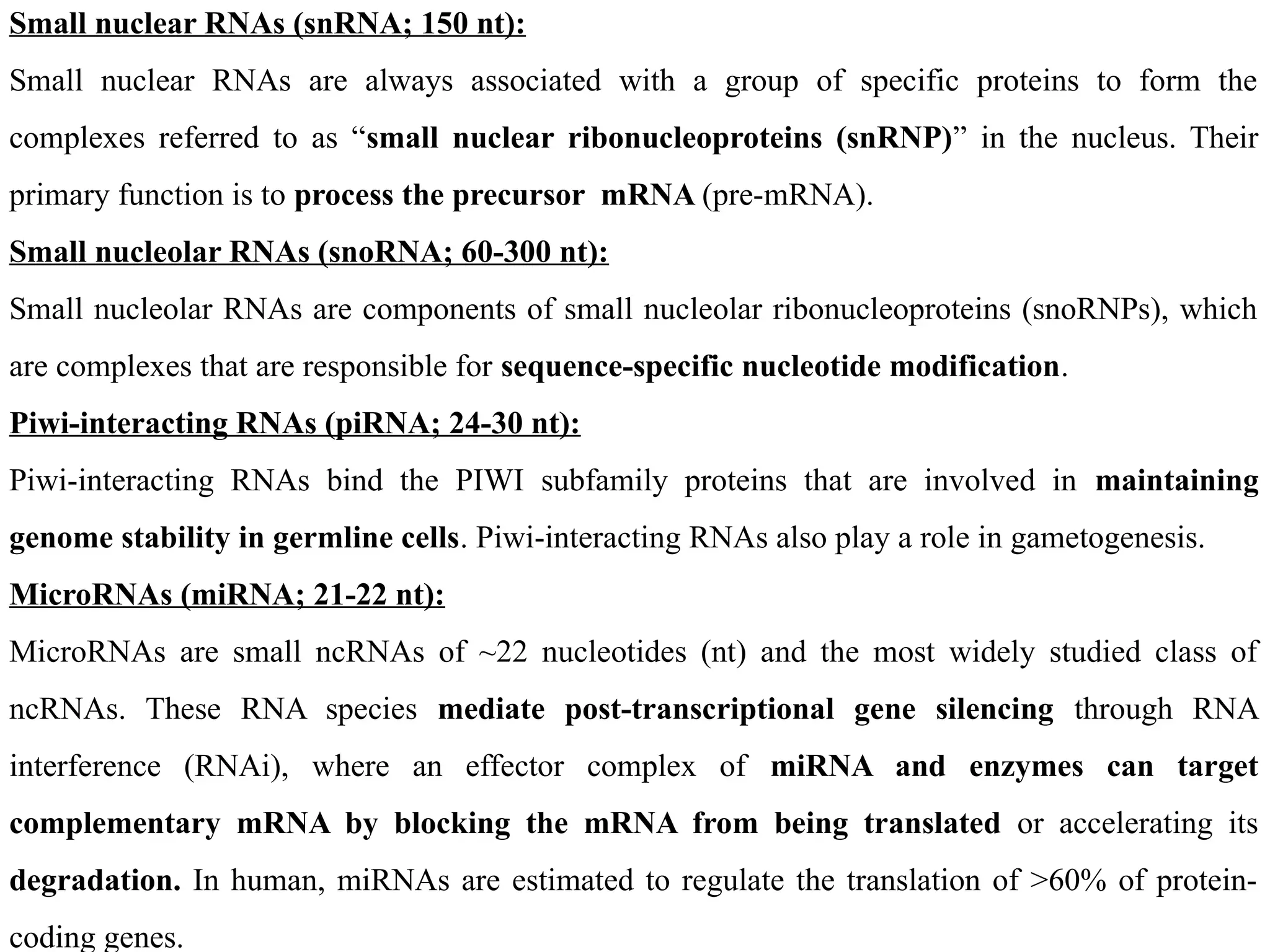
![Prokaryotic Ribosomes Are Composed of 30S and 50S Subunits
Table: 2 Structural Organization of E.coli Ribosomes
70S [Mass=2520 kD]
30S 50S
16S ribosomal RNA (rRNA) 23S rRNA and 5S rRNA
[21 different proteins] [31 different proteins]
Courtesy: Biochemistry by Reginald H. Garrett, Charles M. Grisham (4th ed.)
Page No. 965](https://image.slidesharecdn.com/typesofrna-241113092306-9d01579c/75/Types-of-RNA-structure-and-its-functions-pptx-4-2048.jpg)