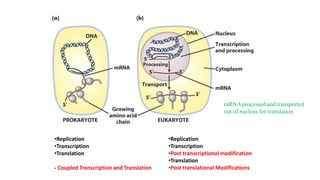
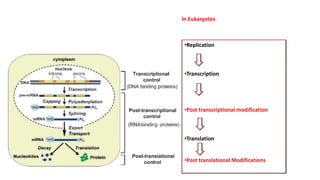
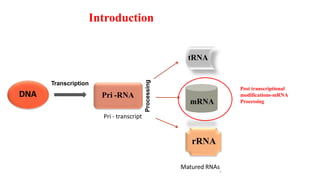
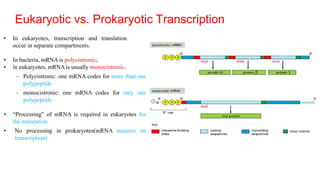
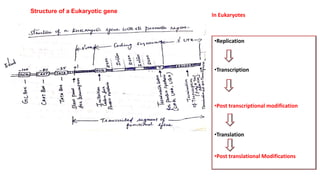
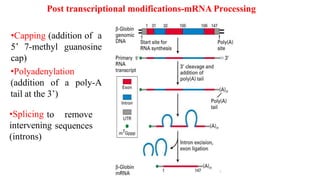
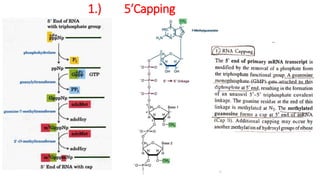
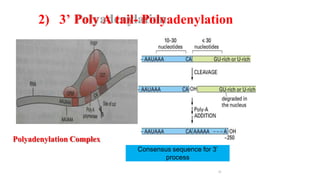
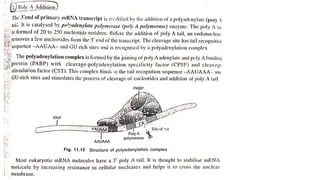
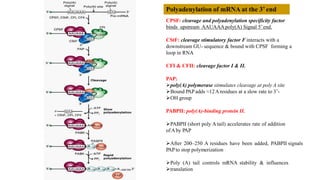
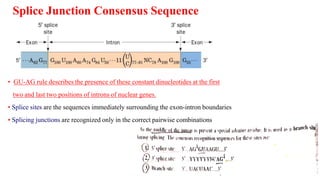
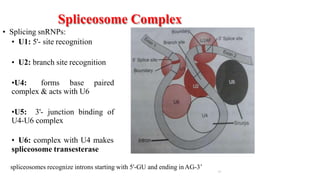
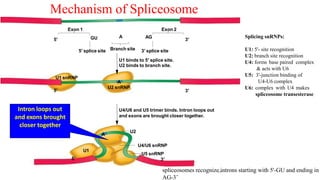
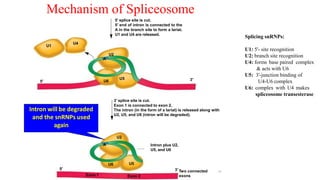
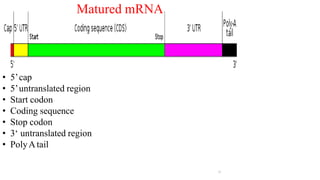
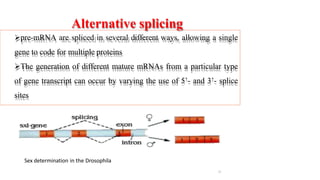
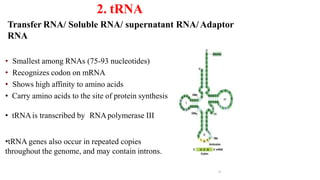
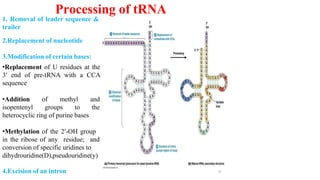
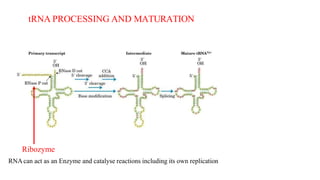
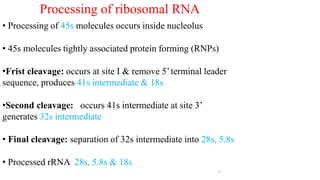
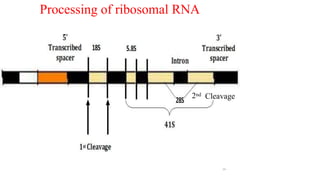
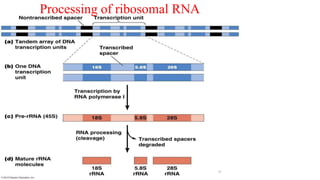
The document discusses post-transcriptional modifications that occur in eukaryotic mRNA. It describes three major modifications: 1) 5' capping, which adds a 7-methylguanosine cap to protect the mRNA and enhance translation, 2) 3' polyadenylation, which adds a poly-A tail to stabilize the mRNA and influence translation and stability, and 3) splicing, by which introns are removed from pre-mRNA through the spliceosome complex to produce mature mRNA for translation.