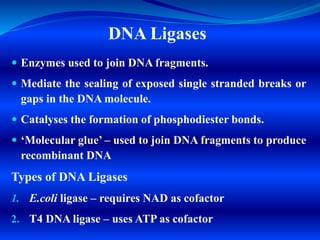
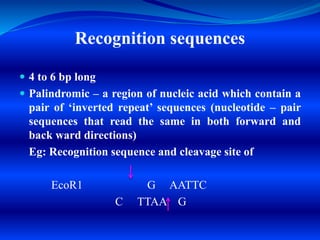
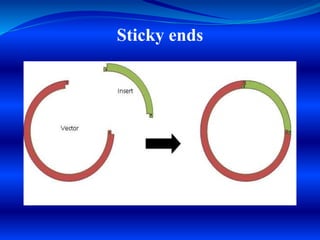
Restriction endonucleases are enzymes found in bacteria that recognize specific nucleotide sequences in DNA and cut the DNA at those sites. This acts as a defense mechanism for bacteria by destroying foreign DNA, such as that from viruses. There are several types of restriction endonucleases, but type II are most useful for genetic engineering as they cut DNA at predictable, site-specific locations. The resulting DNA fragments can be joined back together via DNA ligases. Restriction endonucleases have four- to six-letter recognition sequences that are palindromic, and they produce sticky or blunt ends depending on whether cuts are offset or at the same position.






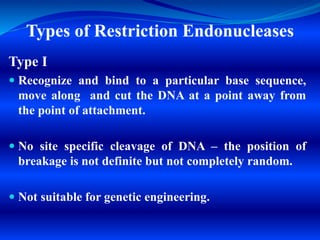
![Type II
Site specific cleavage – recognize a specific
sequence in DNA and cut at the same site.
Used in Genetic Engineering
Type III
No site specific cleavage – cut the DNA at a specific
point that is a measured distance (about 25 bp
[bp= base pair] from the recognition sequence)
Not suitable for Genetic Engineering](https://image.slidesharecdn.com/restrictionendonucleasesdnaligase-200327150304/85/RESTRICTION-ENDONUCLEASES-DNA-LIGASES-SMG-10-320.jpg)