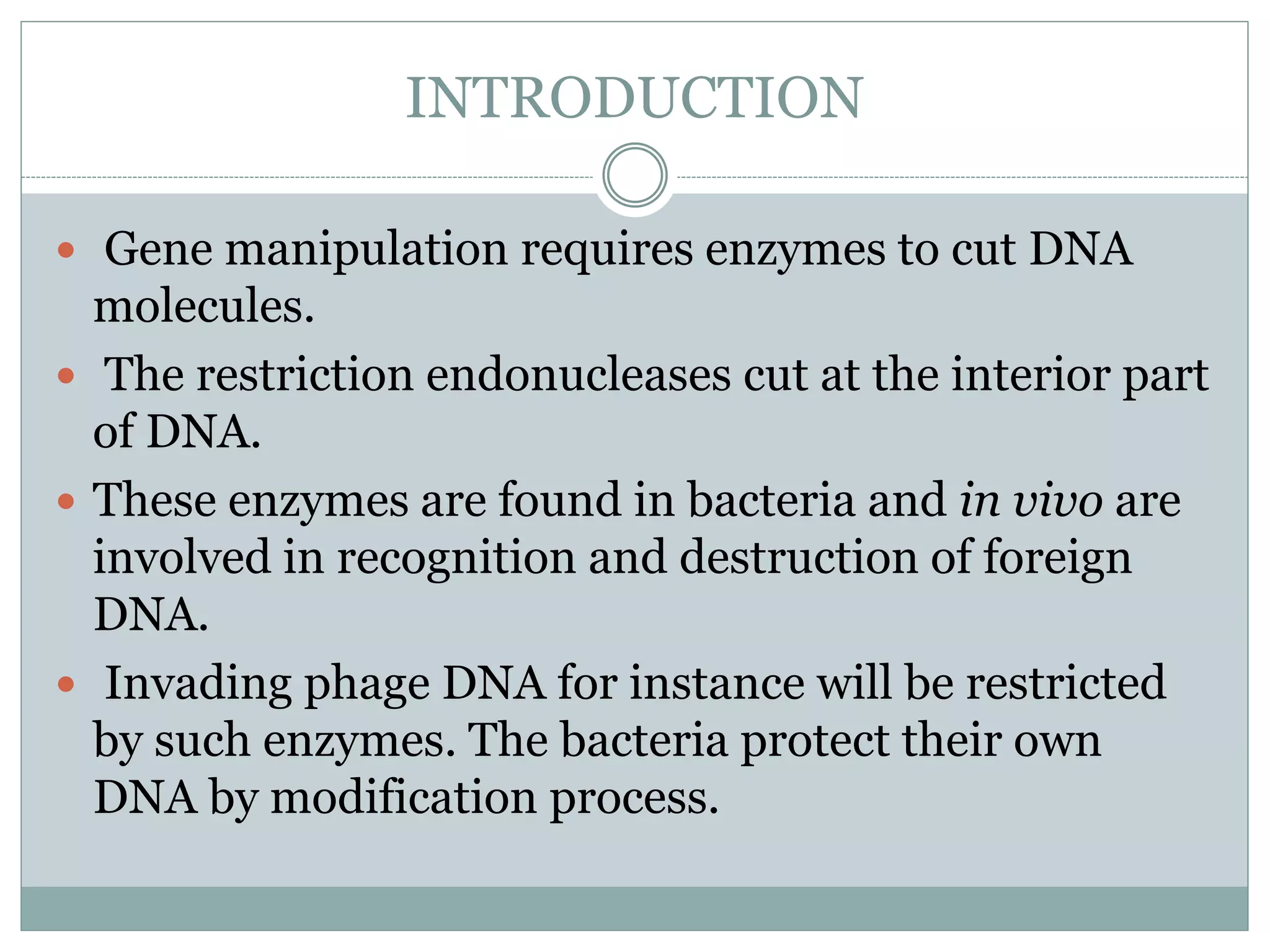
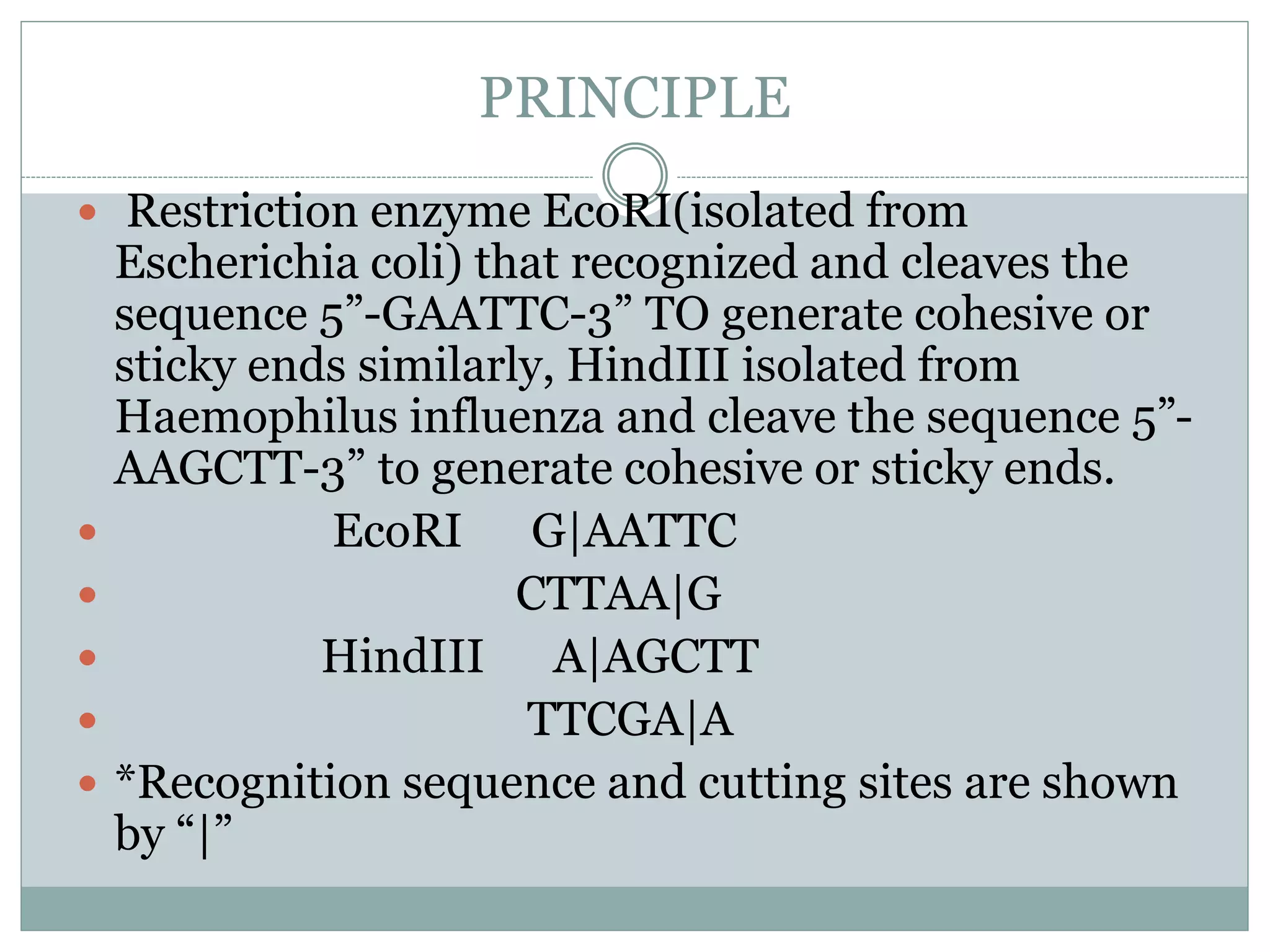
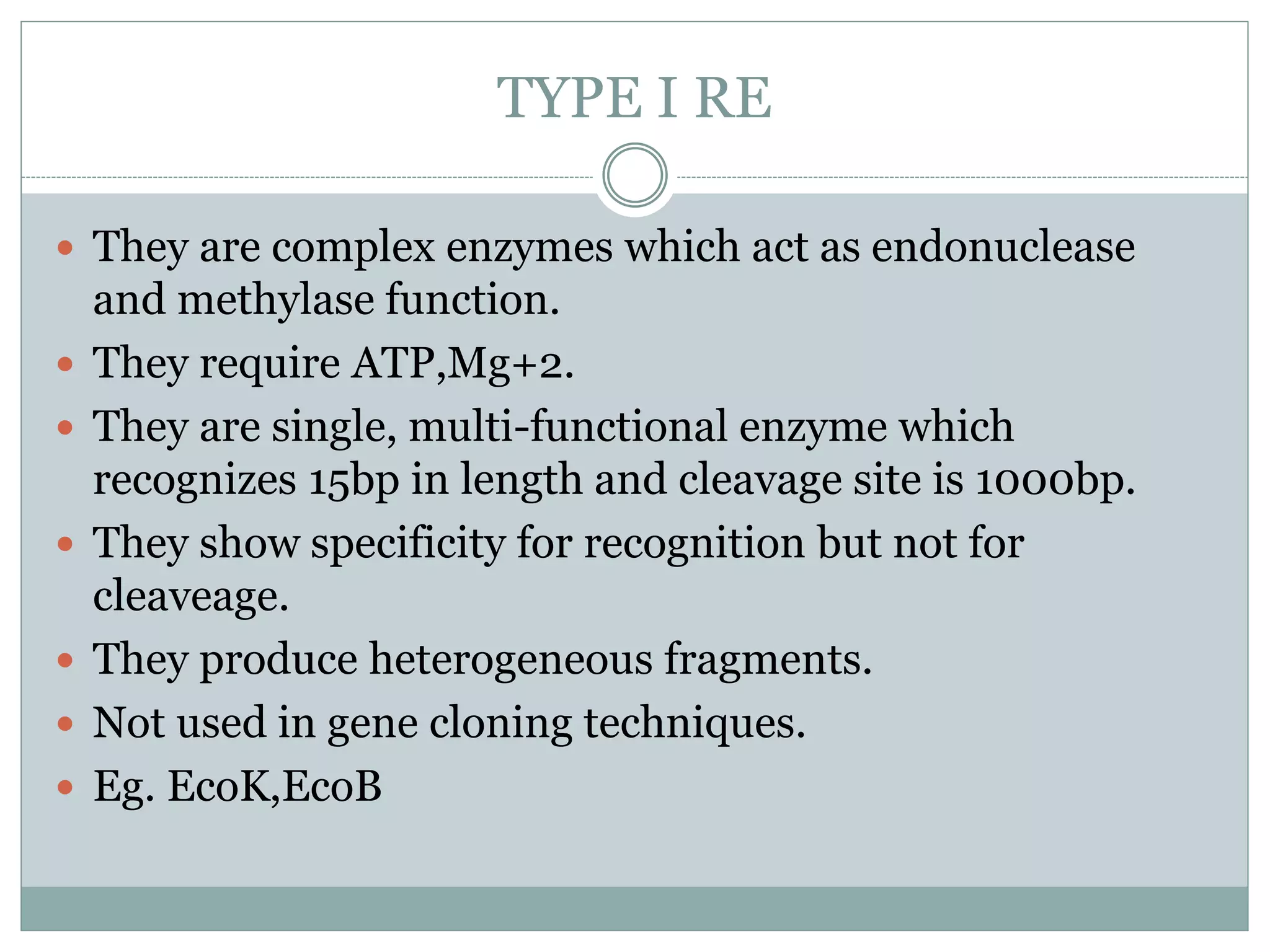
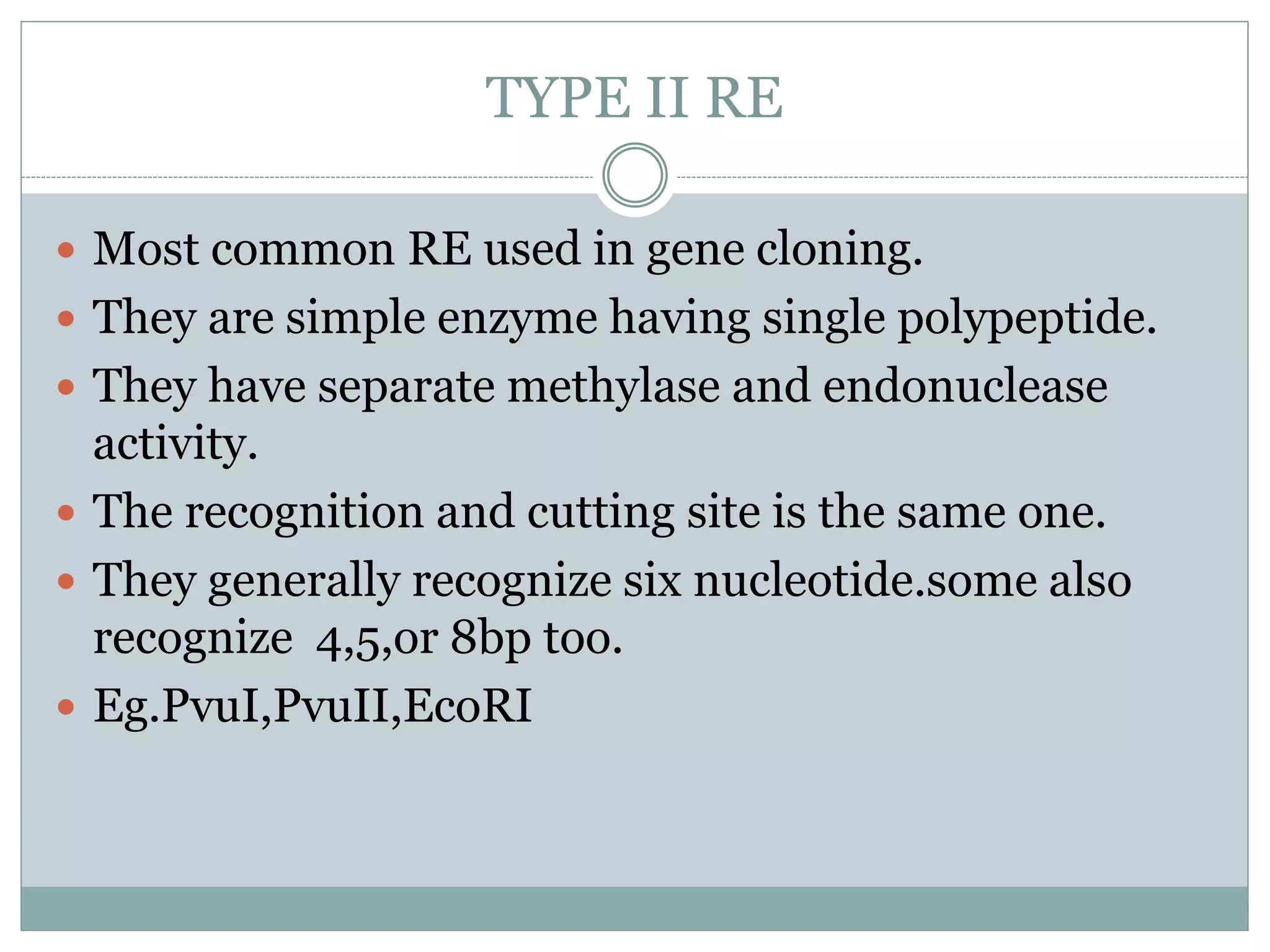
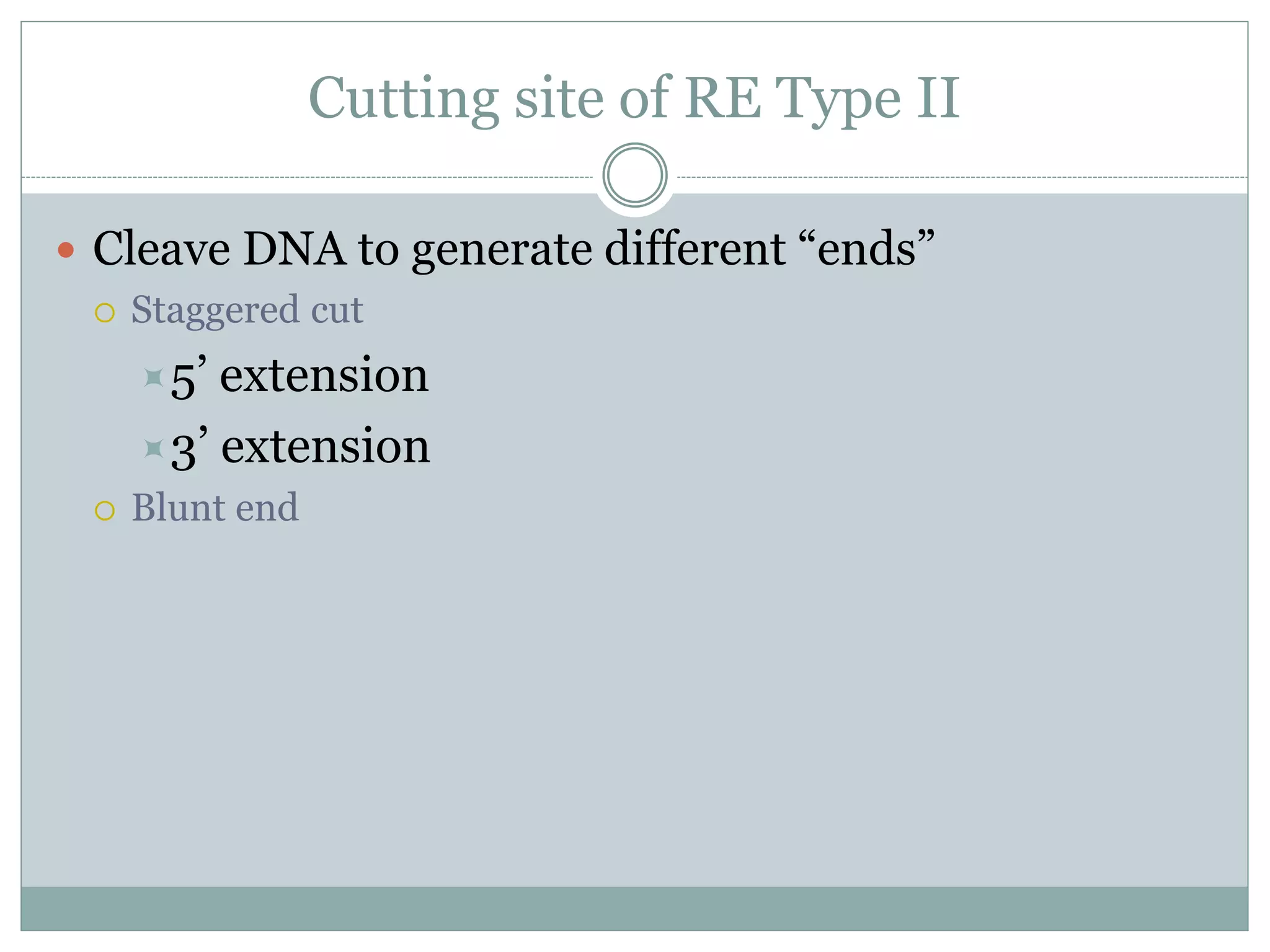
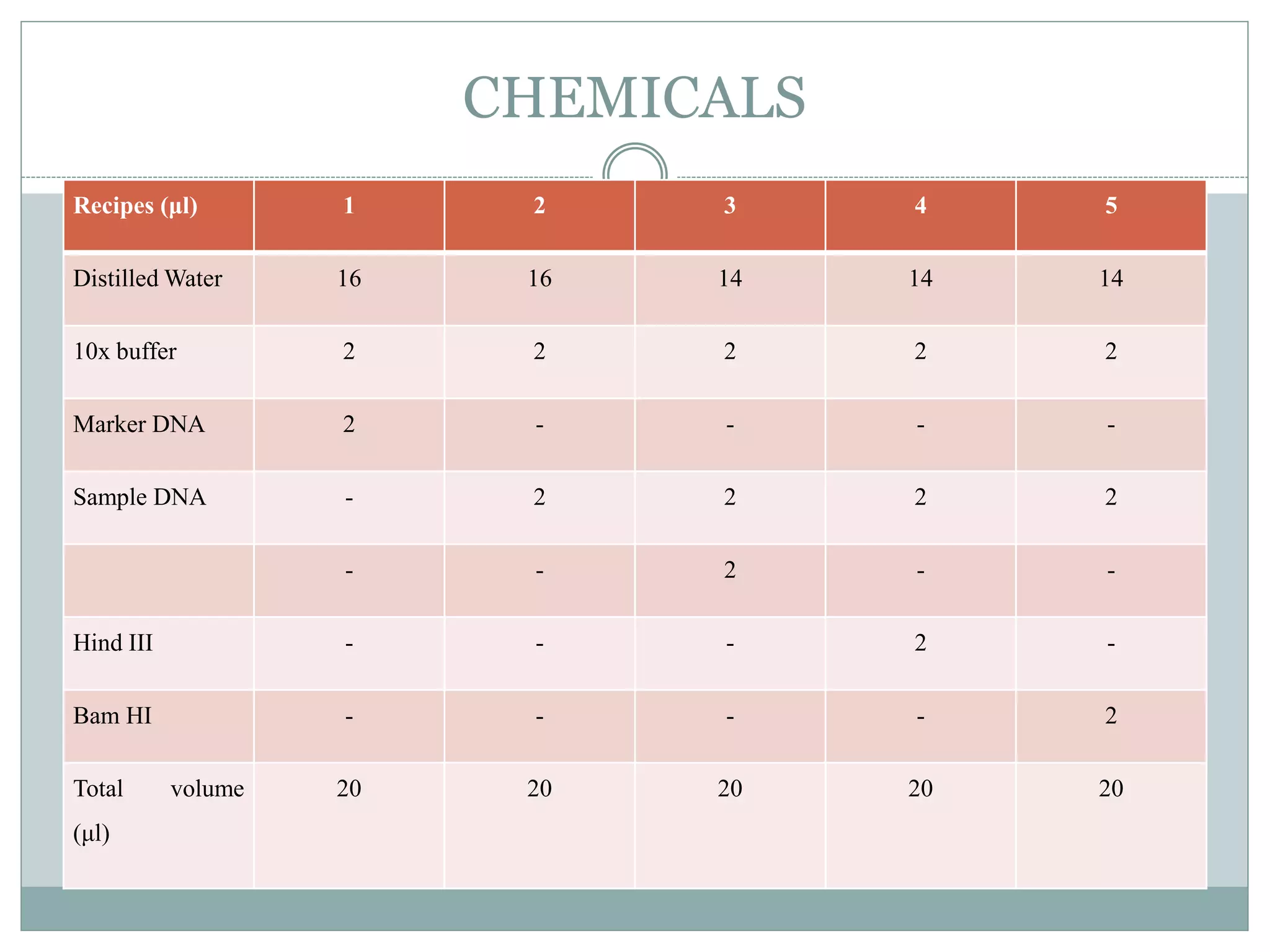
Restriction enzymes are enzymes found in bacteria that cut DNA molecules at specific recognition sequences. They help with gene manipulation by cutting DNA at targeted locations. There are three main types of restriction enzymes: Type I recognize long sequences and cut elsewhere; Type II recognize short palindromic sequences and cut within; Type III have complex recognition sites and cutting. Restriction enzyme digestion is used in techniques like cloning and diagnostic restriction mapping by targeting specific sequences for cleavage.