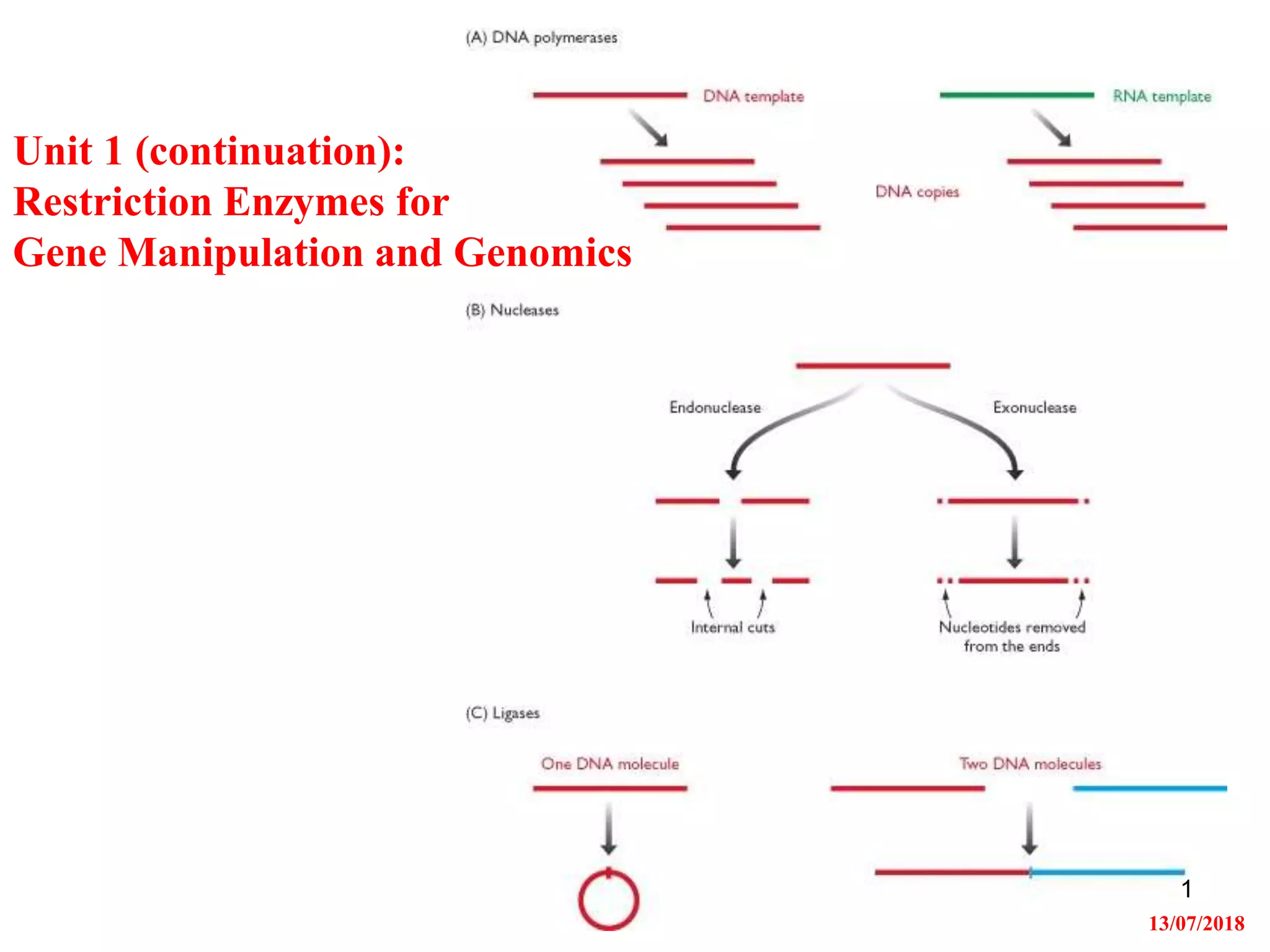
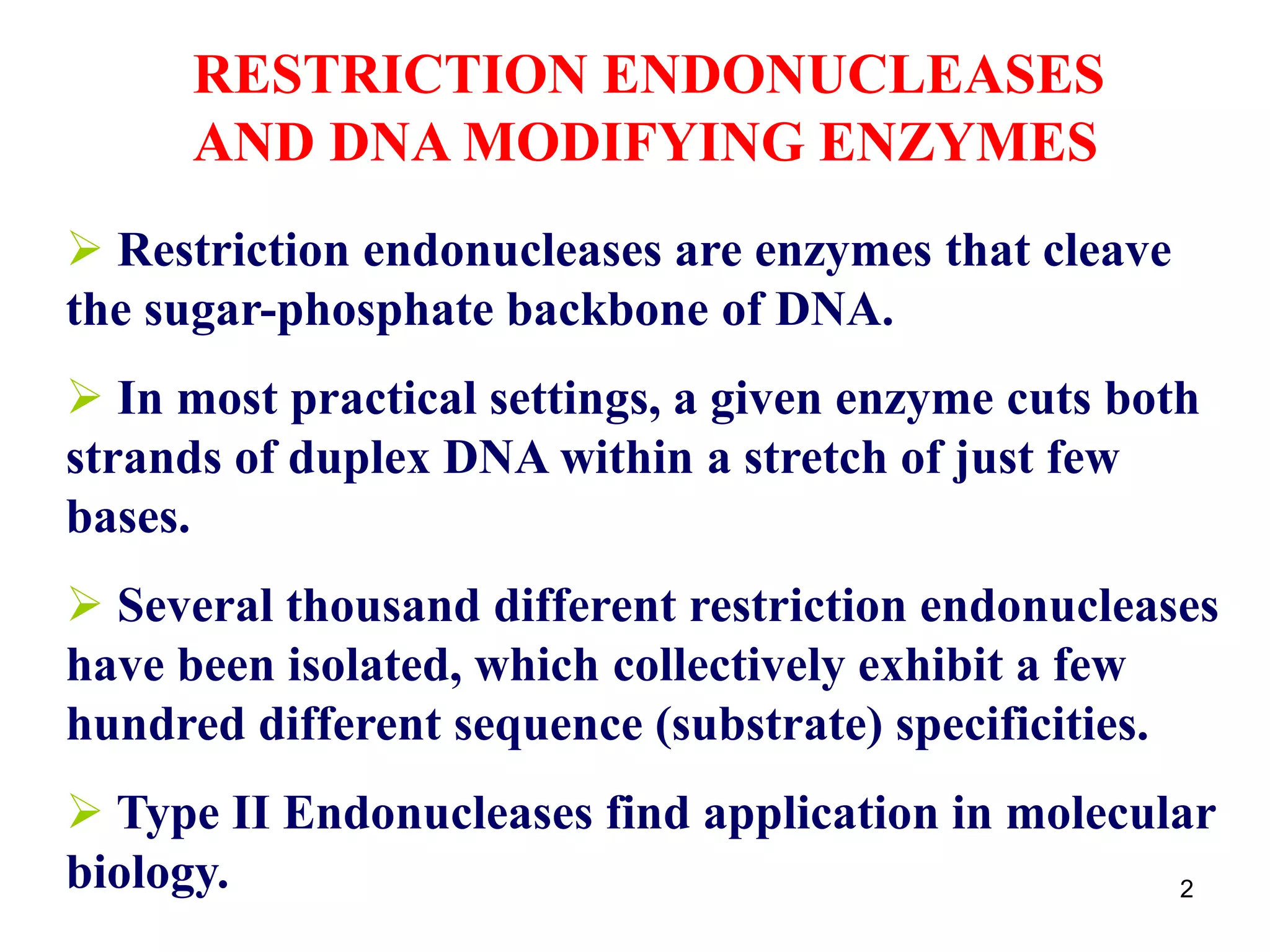
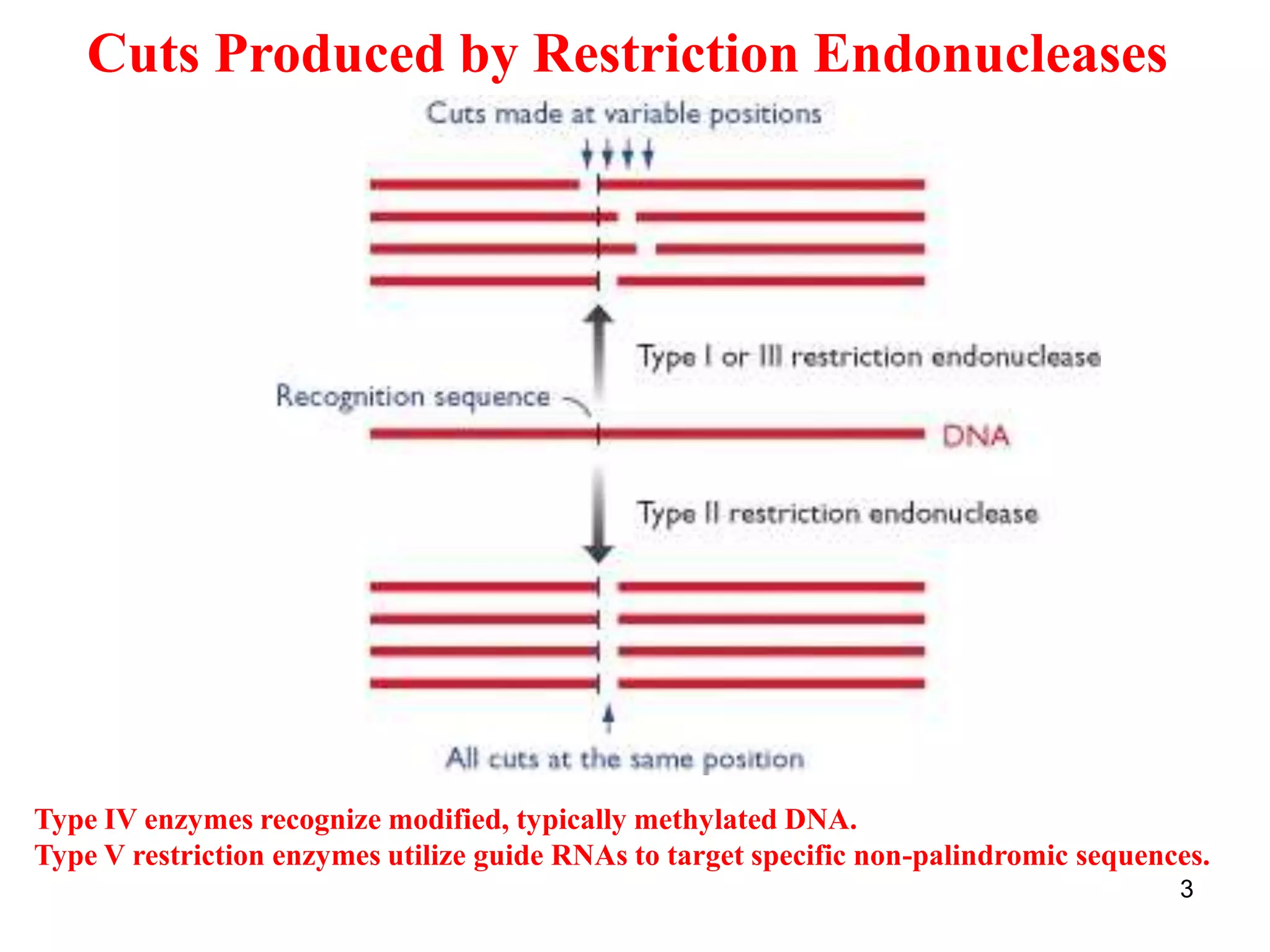
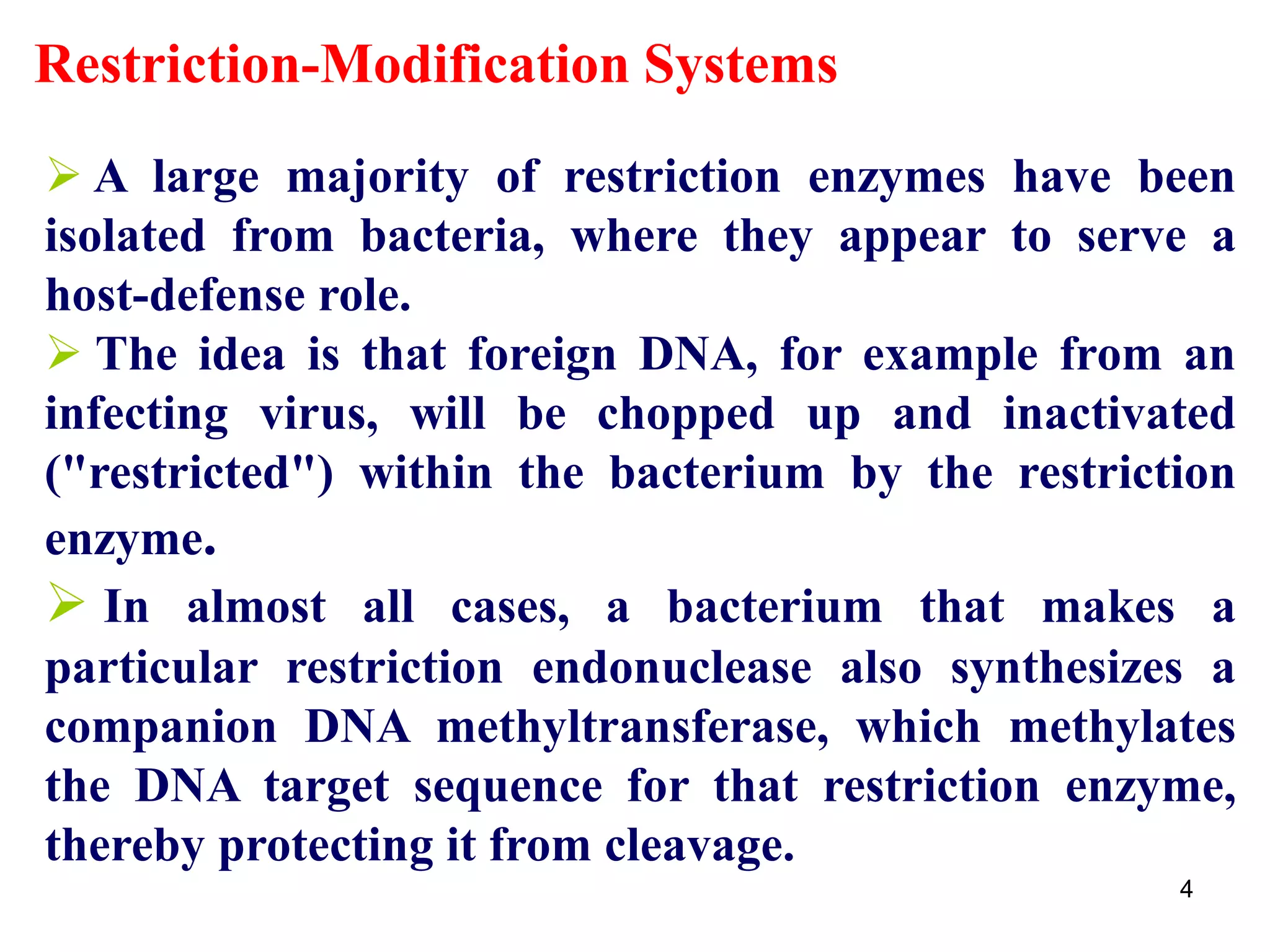
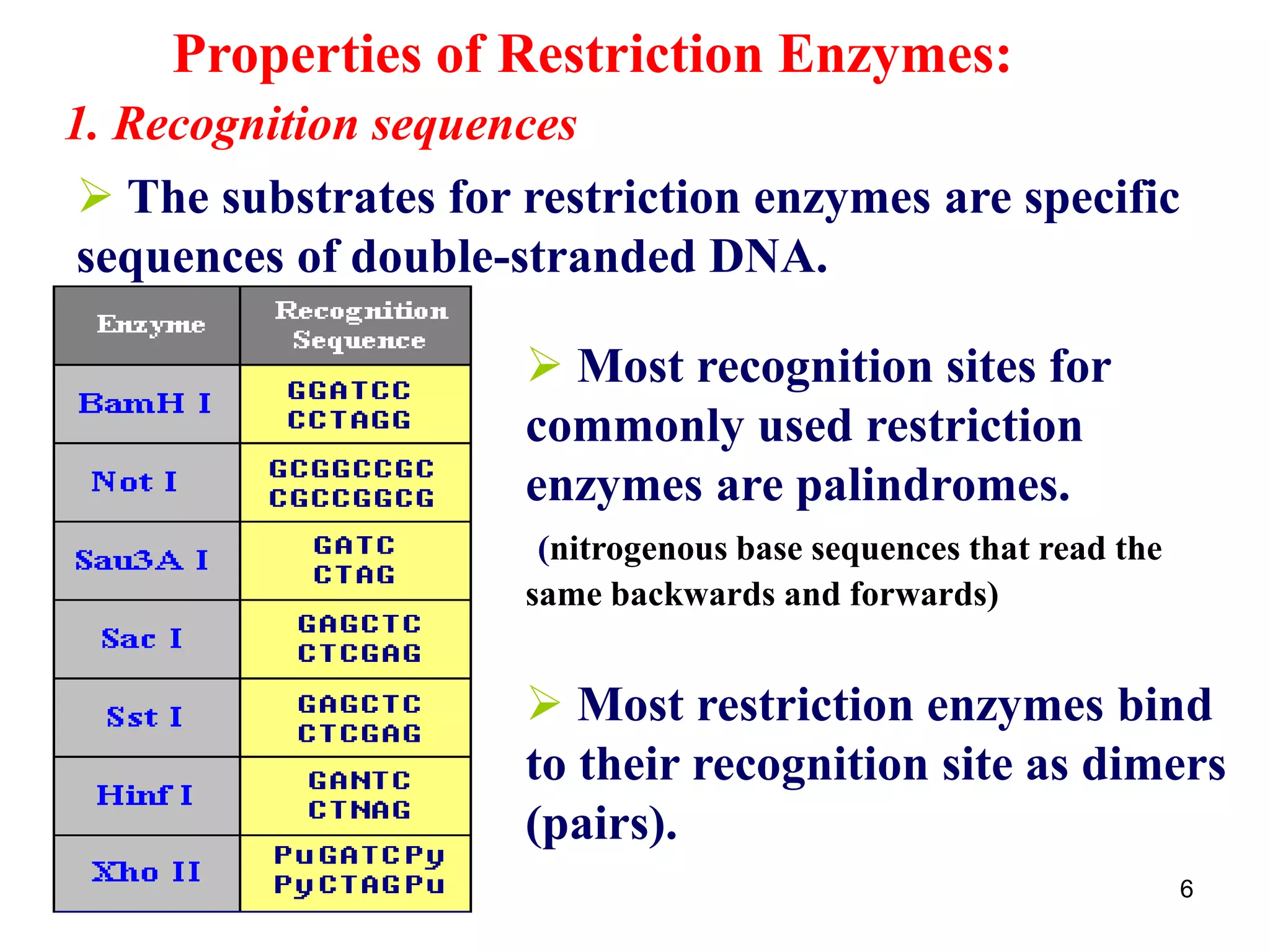
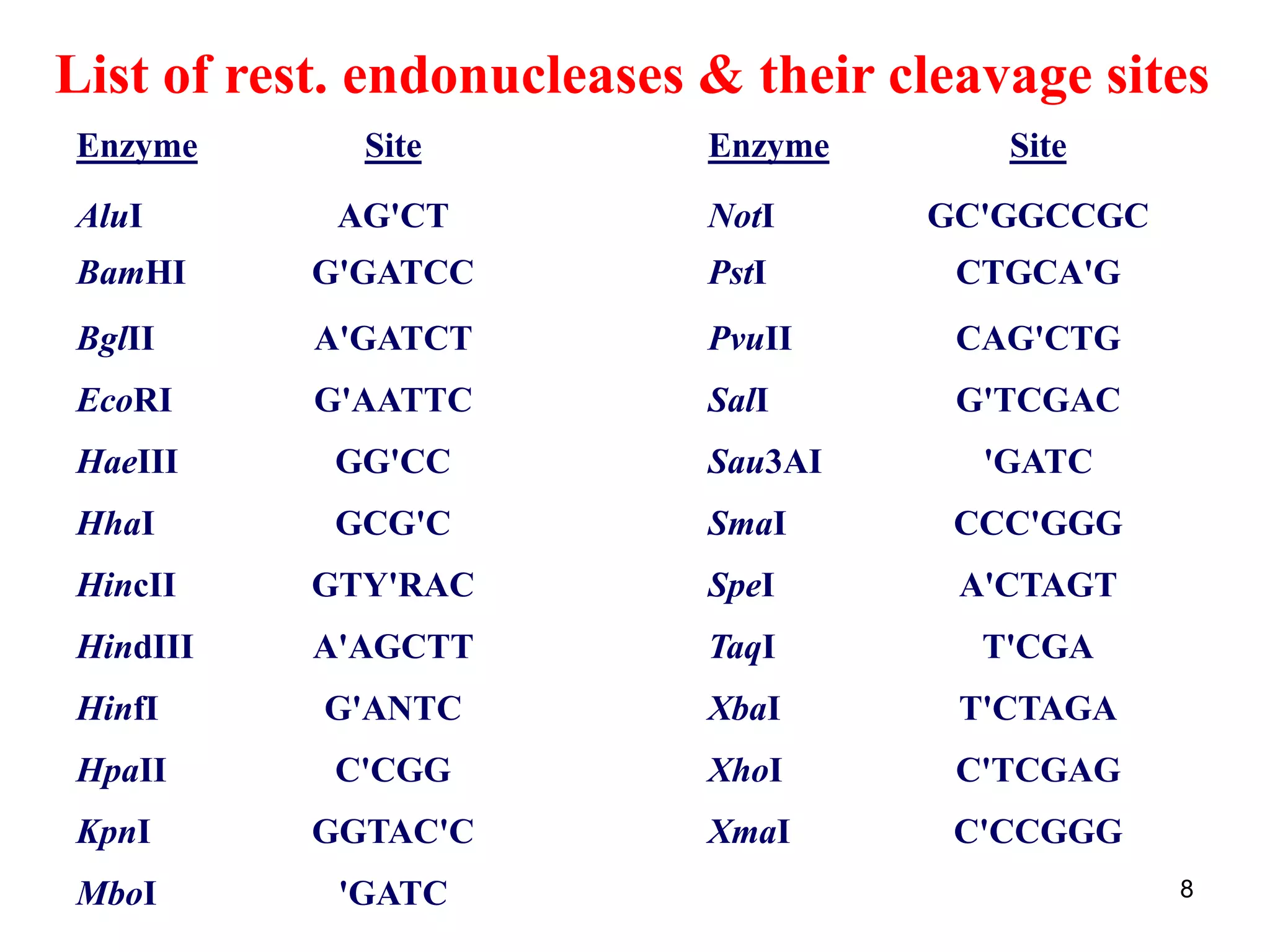
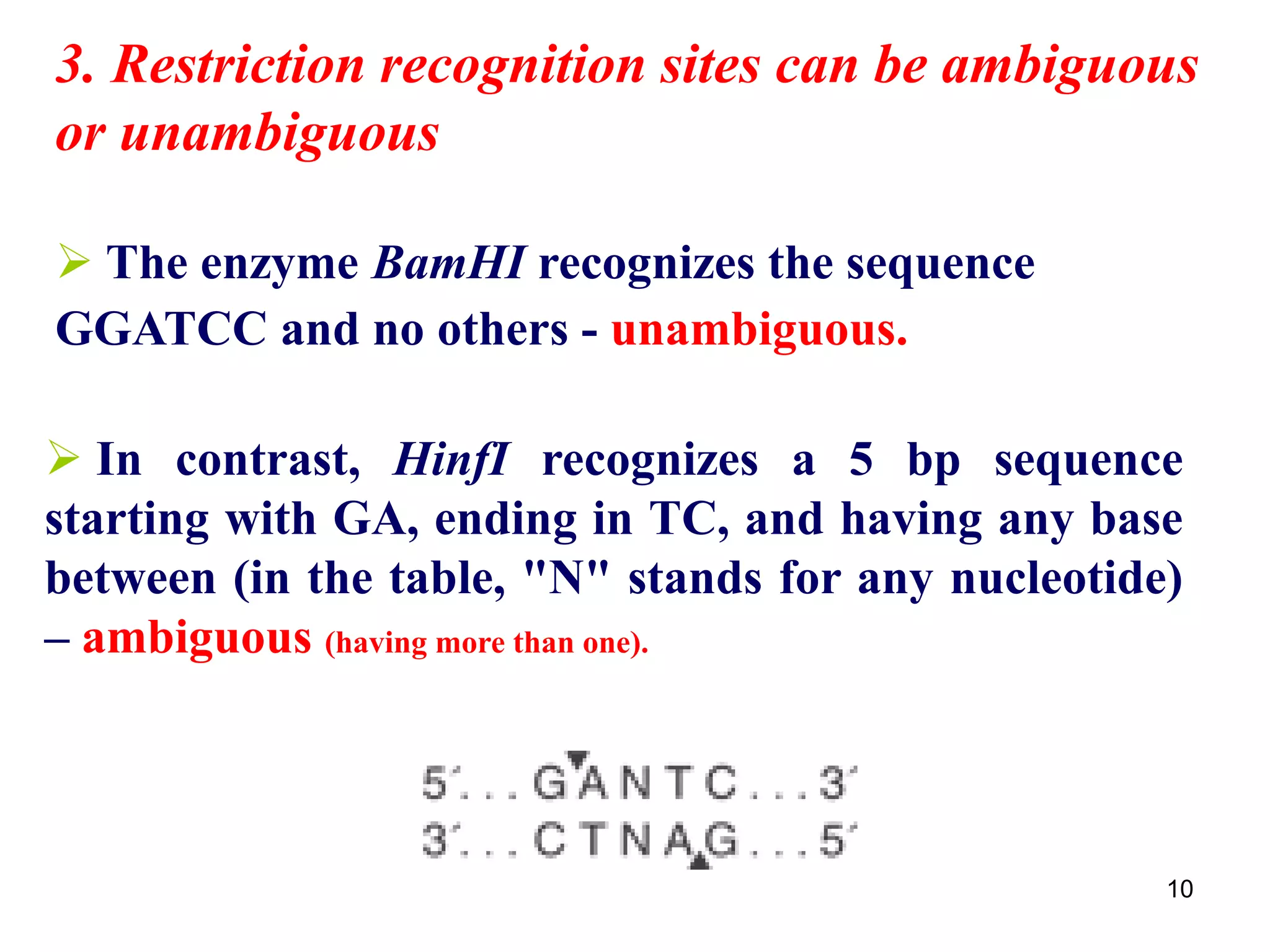
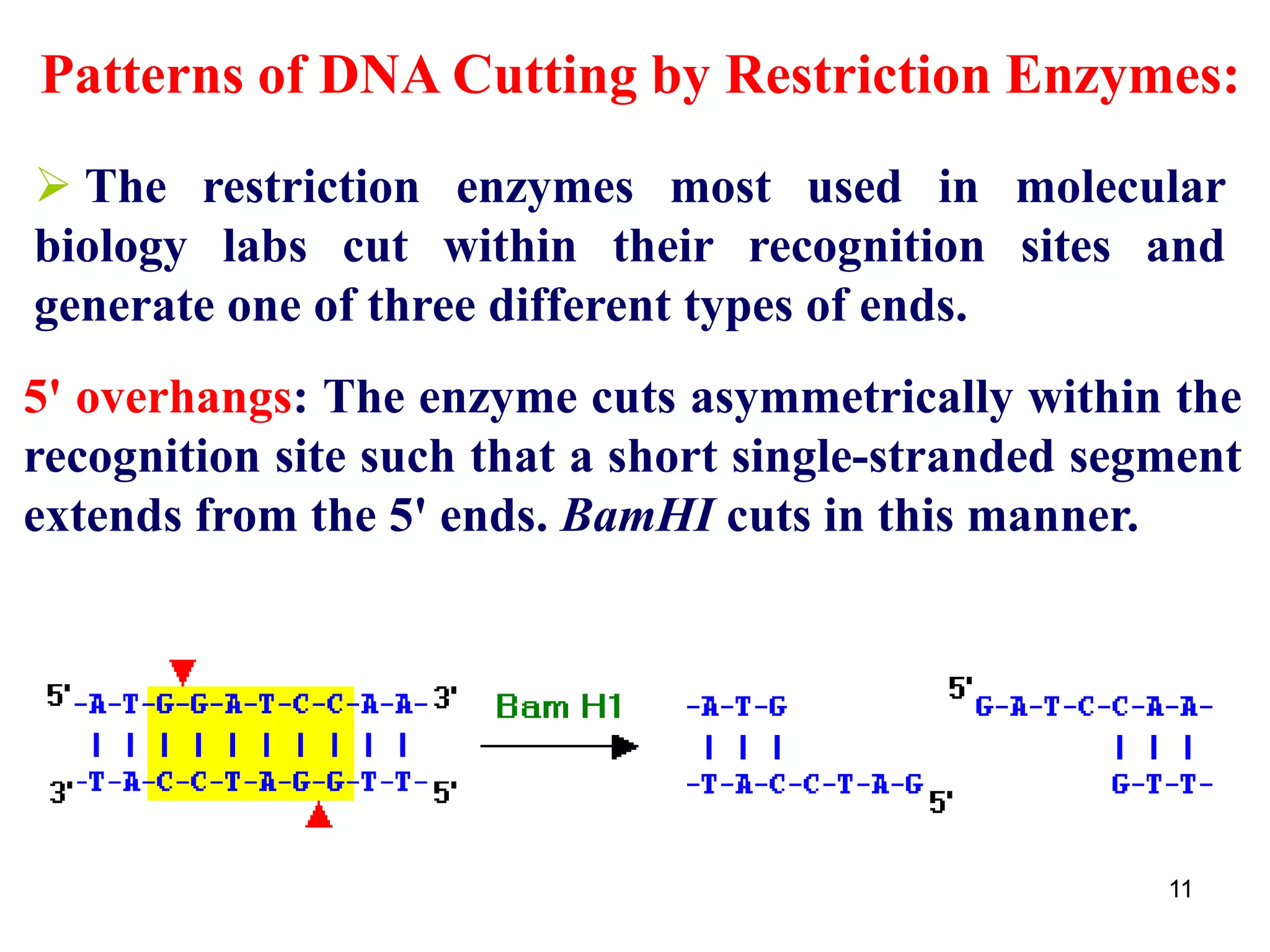
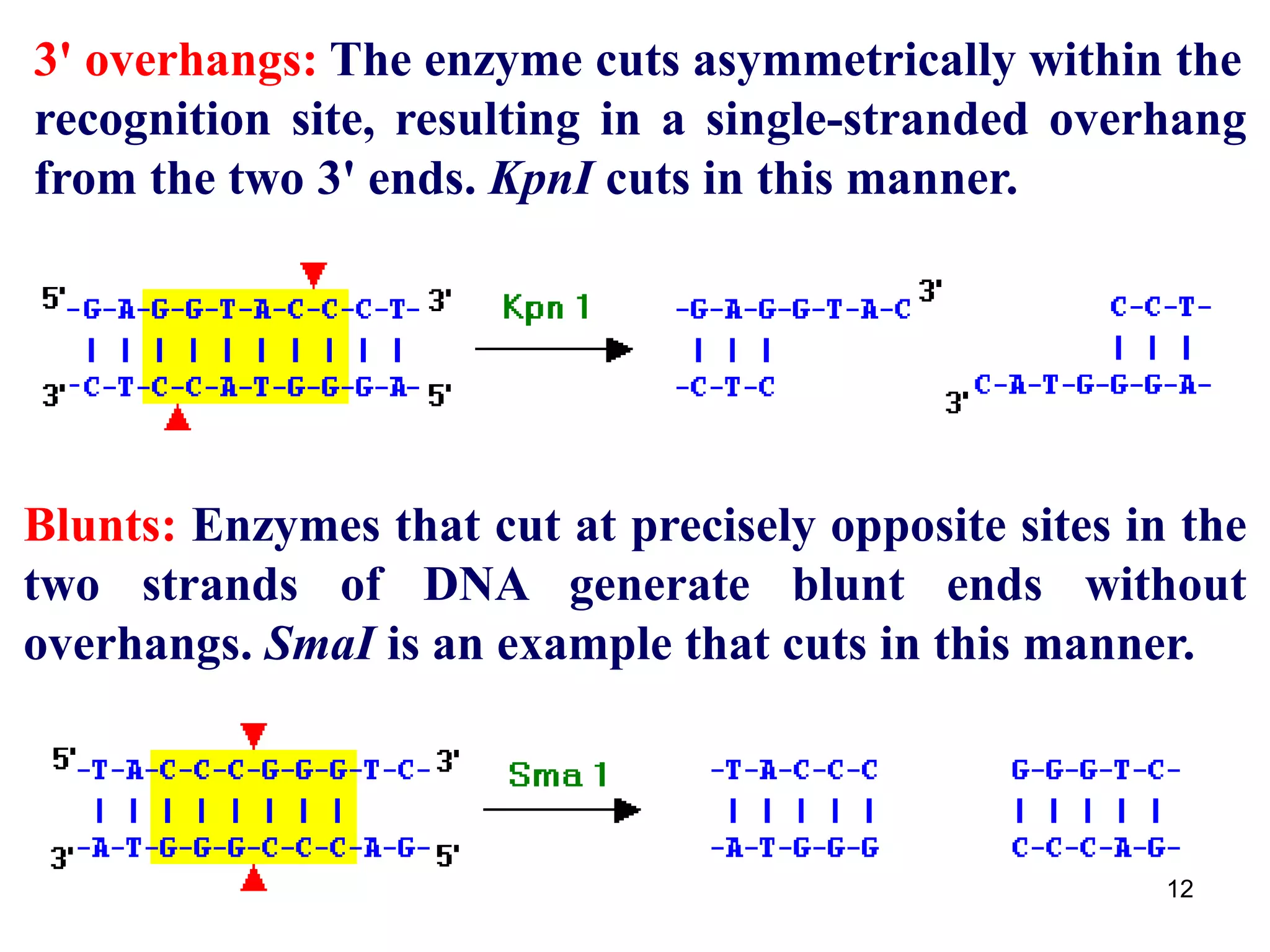
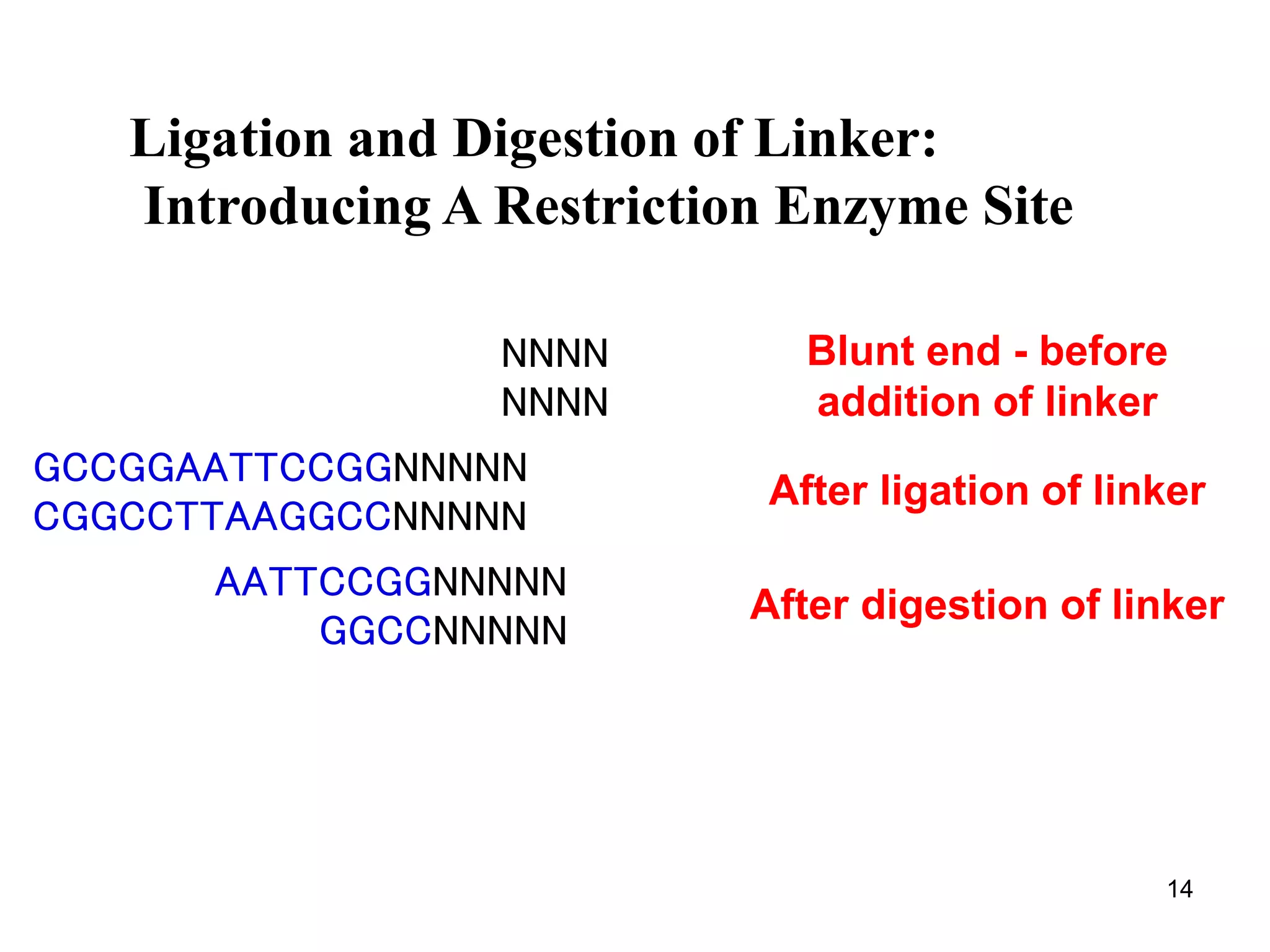
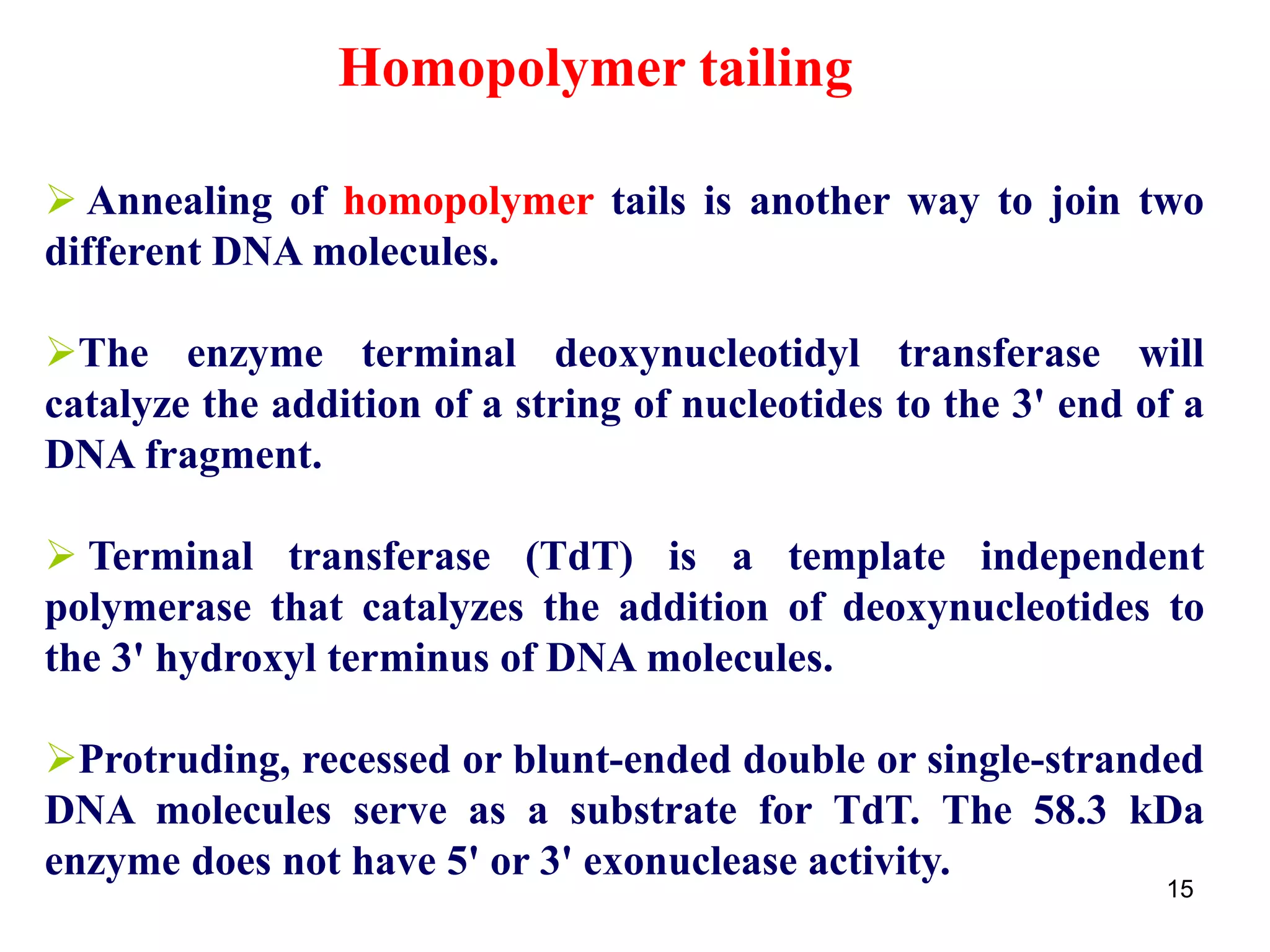
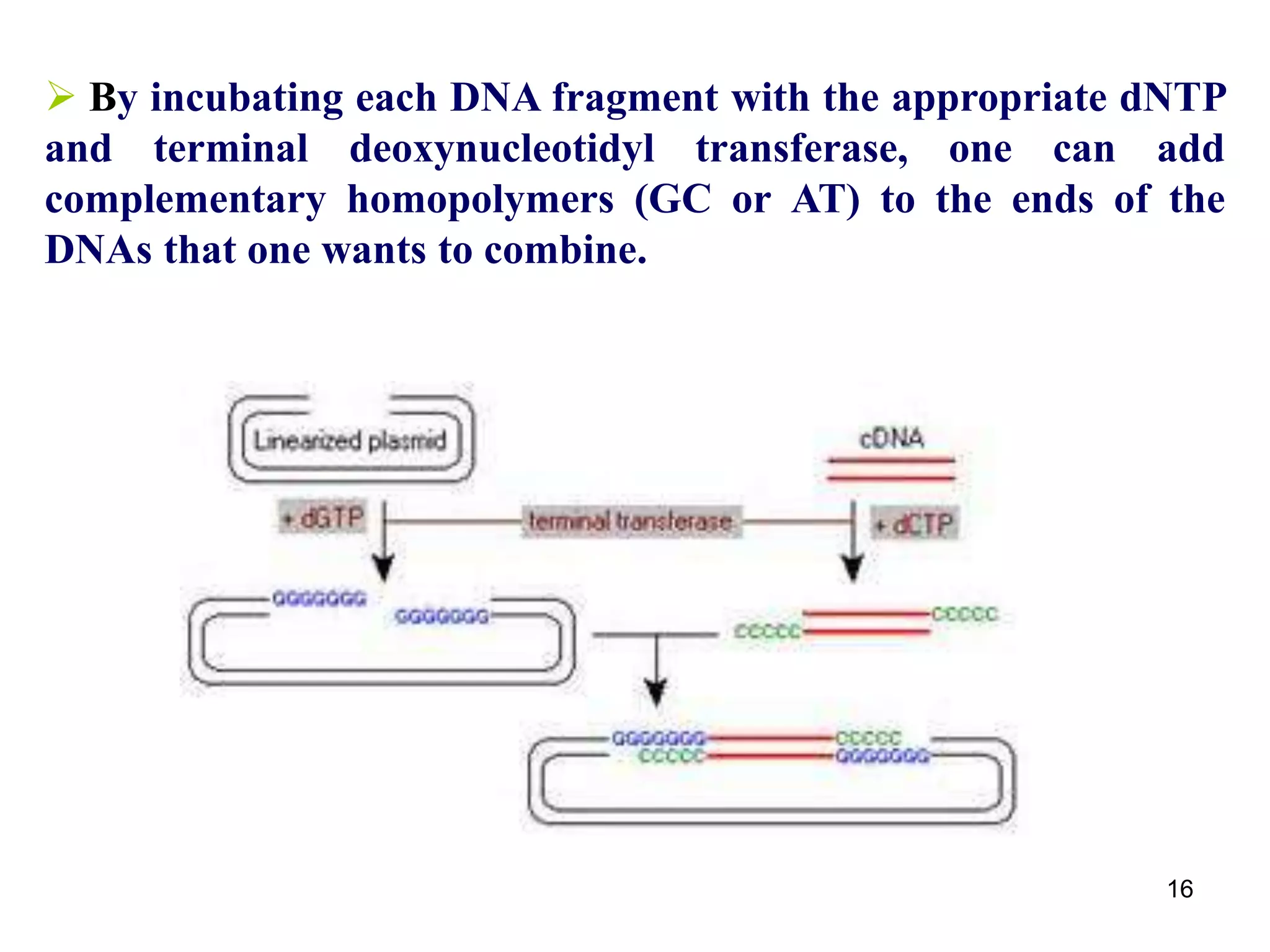
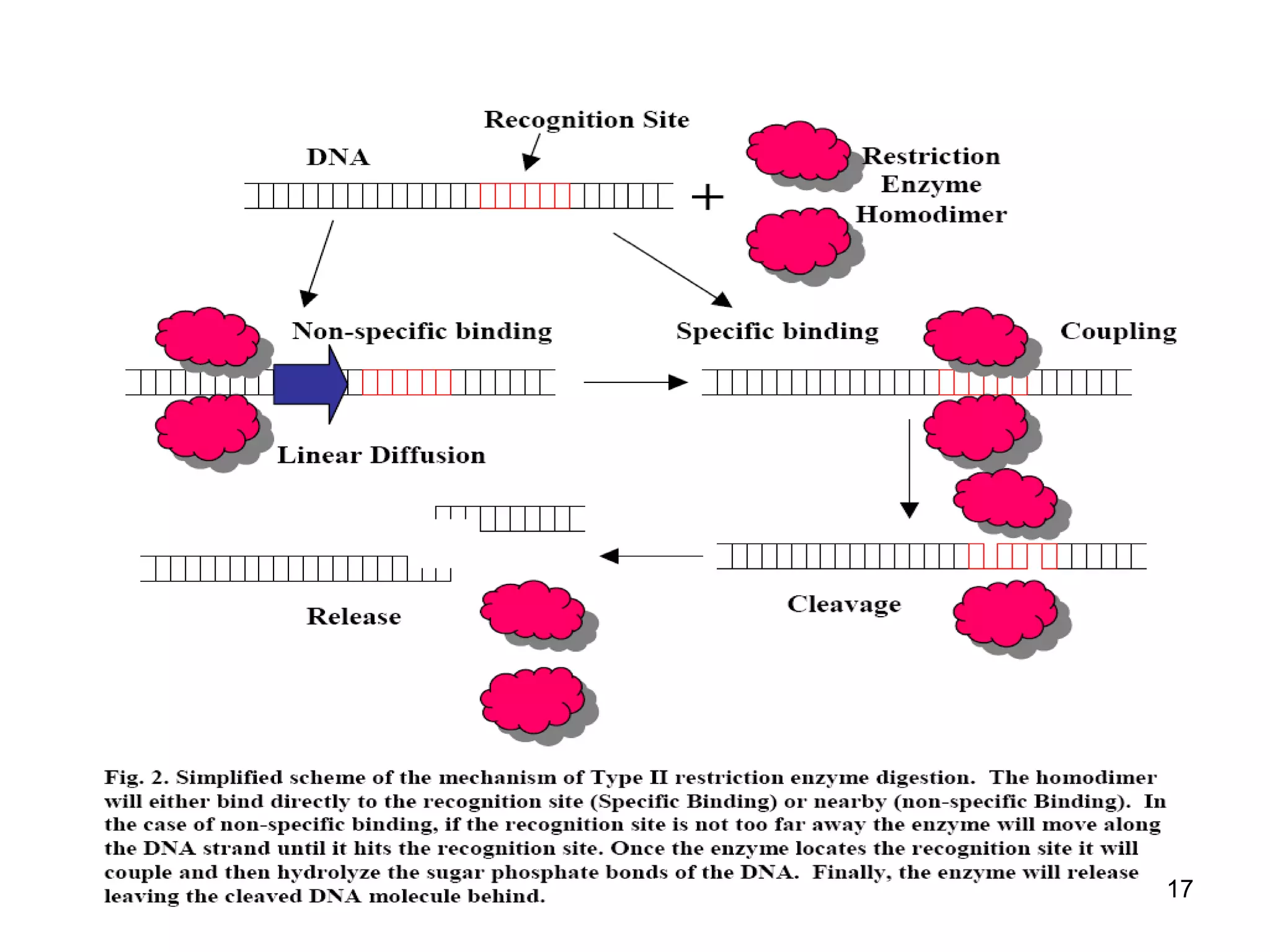
Restriction endonucleases are enzymes that cut DNA at specific recognition sequences. There are several thousand restriction enzymes that recognize different DNA sequences. Most are isolated from bacteria and serve to defend the host by cutting foreign DNA. Each restriction enzyme requires a specific recognition sequence, which can be 4-8 base pairs long. They cut DNA, leaving either blunt ends, 5' overhangs, or 3' overhangs. Linkers or homopolymer tails can be added to DNA fragments to allow them to be joined together using restriction sites or base pairing. Restriction enzymes are important tools in molecular cloning and genomics.