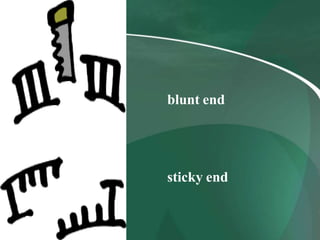
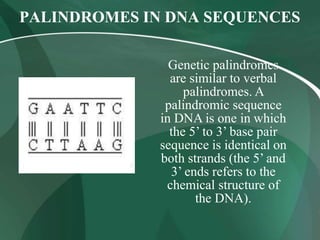
Restriction enzymes are proteins that cut DNA at specific recognition sequences. They are classified into three types based on their composition and target site. Type I enzymes recognize asymmetric sites and require cofactors. Type II enzymes recognize palindromic sites and require magnesium. Type III enzymes recognize two non-palindromic sites and require cofactors. Restriction enzymes are widely used in gene cloning, DNA manipulation, and other applications that involve modifying DNA.