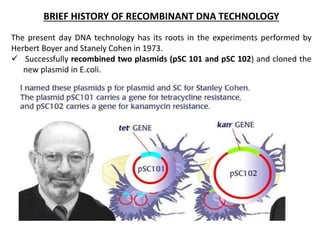
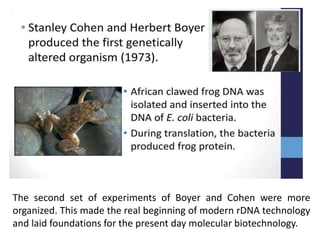
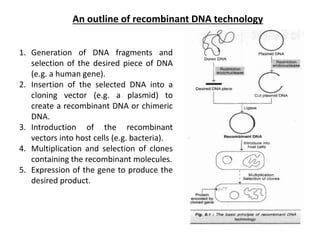
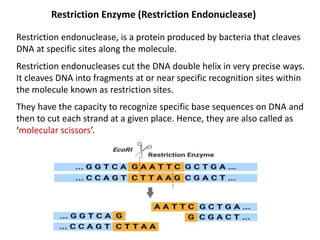
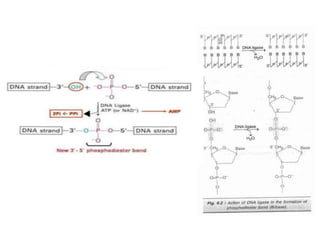
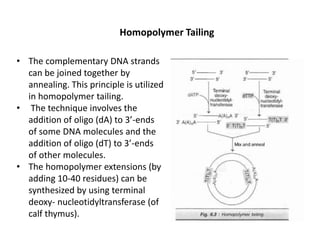
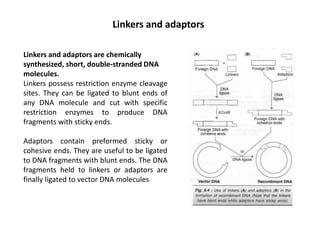
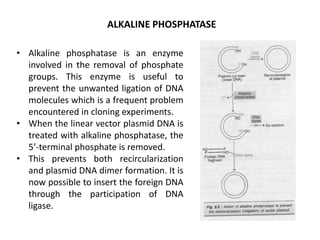
This document discusses the key enzymes involved in recombinant DNA technology. It describes how restriction enzymes cut DNA at specific recognition sites, and DNA ligases join cut DNA fragments back together. The document outlines the process of recombinant DNA technology, including generating DNA fragments, inserting them into cloning vectors, introducing the vectors into host cells, and expressing the gene of interest. It provides details on various restriction enzymes and DNA-modifying enzymes used in genetic engineering applications.