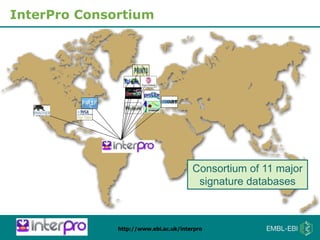
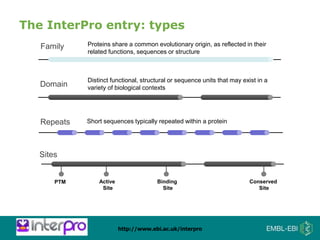
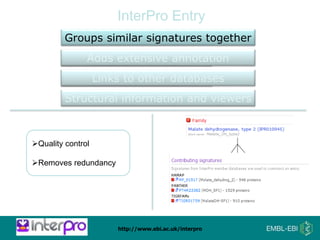
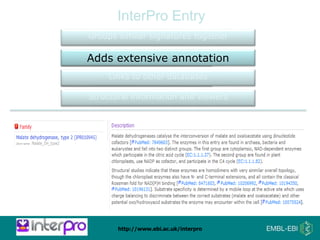
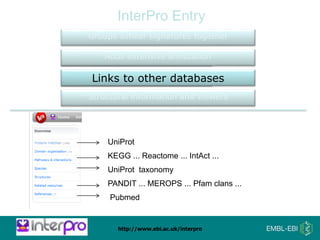
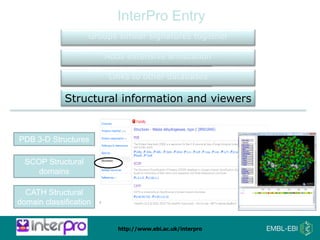
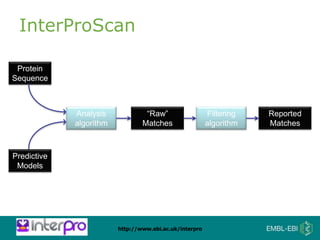
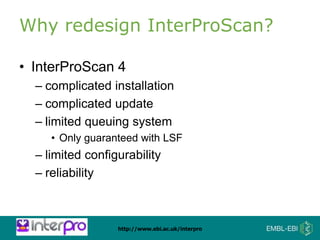
The document provides an overview of InterPro, a database from the European Molecular Biology Laboratory that groups predictive protein signatures from 11 member databases for functional protein analysis. It discusses the significance of predictive annotation tools, the structures involved in the analysis of proteins, and highlights features of the latest tool, InterProScan 5, including its ease of use and flexible architecture. Additionally, it outlines future functionalities and ongoing developments associated with InterPro and InterProScan.