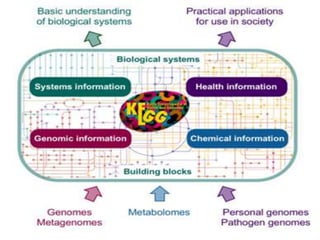
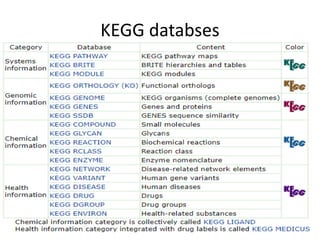
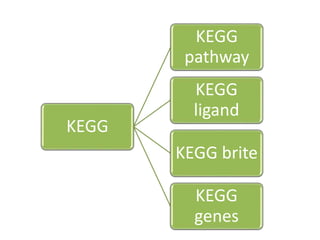
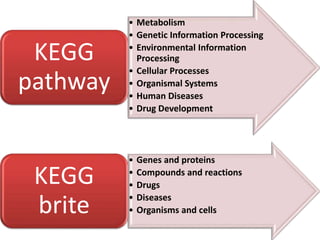
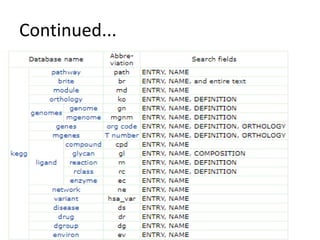
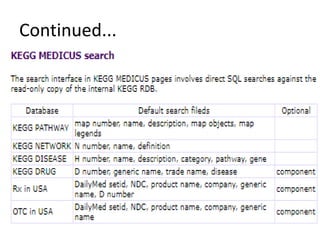
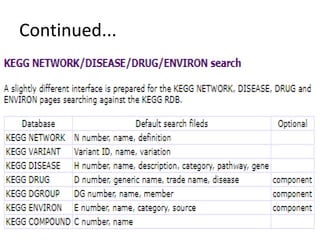
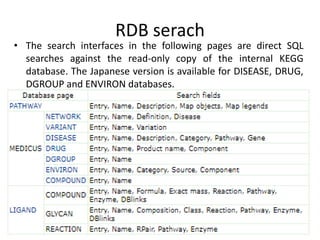
Rashi Srivastava presented on the KEGG database in biotechnology. KEGG is a database that contains genomic, chemical, and systems information to understand biological functions from the molecular level up. It includes pathways, genes, compounds, diseases, drugs, and organisms. KEGG can be searched through its flat file format using DBGET or through its relational database format for more complex queries. It also contains the KEGG MEDICUS search tool and direct SQL searches of its relational database.