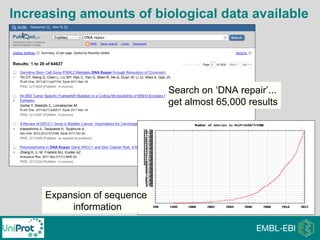
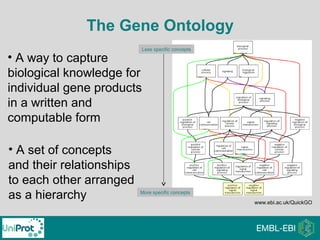
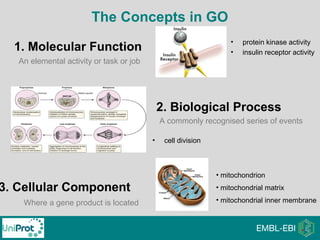
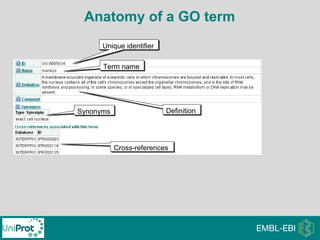
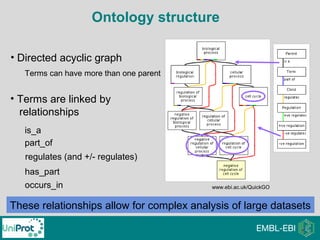
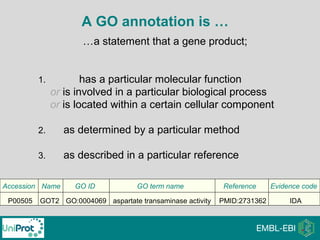
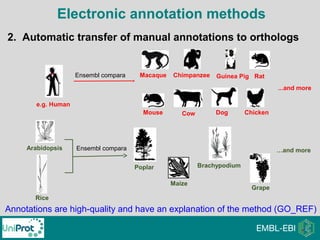
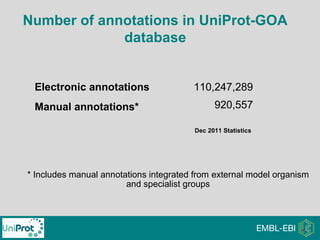
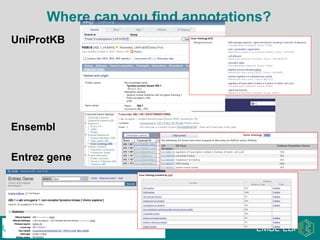
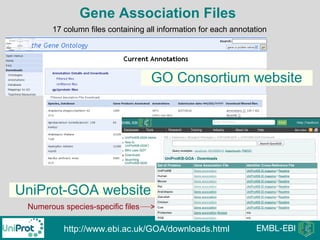
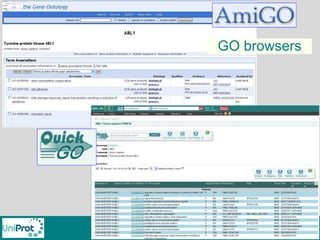
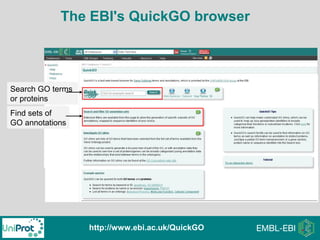
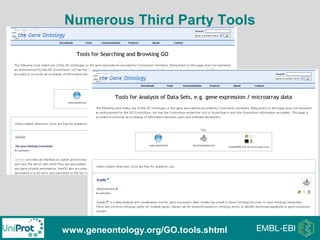
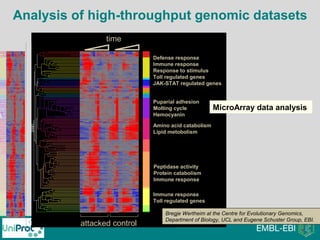
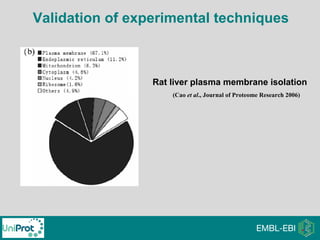
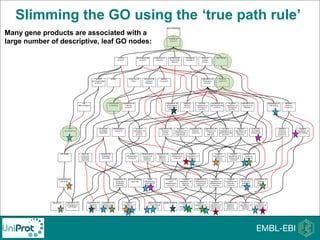
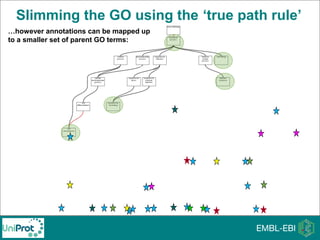
The document provides an introduction to the Gene Ontology (GO) and its annotation resources, emphasizing the need for a standardized vocabulary to address inconsistencies and facilitate comparisons across biological data. It outlines the structure of GO, which consists of molecular functions, biological processes, and cellular components, and details the methods of annotation used to link gene products to these terms. Additionally, it describes how researchers can utilize GO annotations for functional analysis and dataset interpretations through various tools and databases.