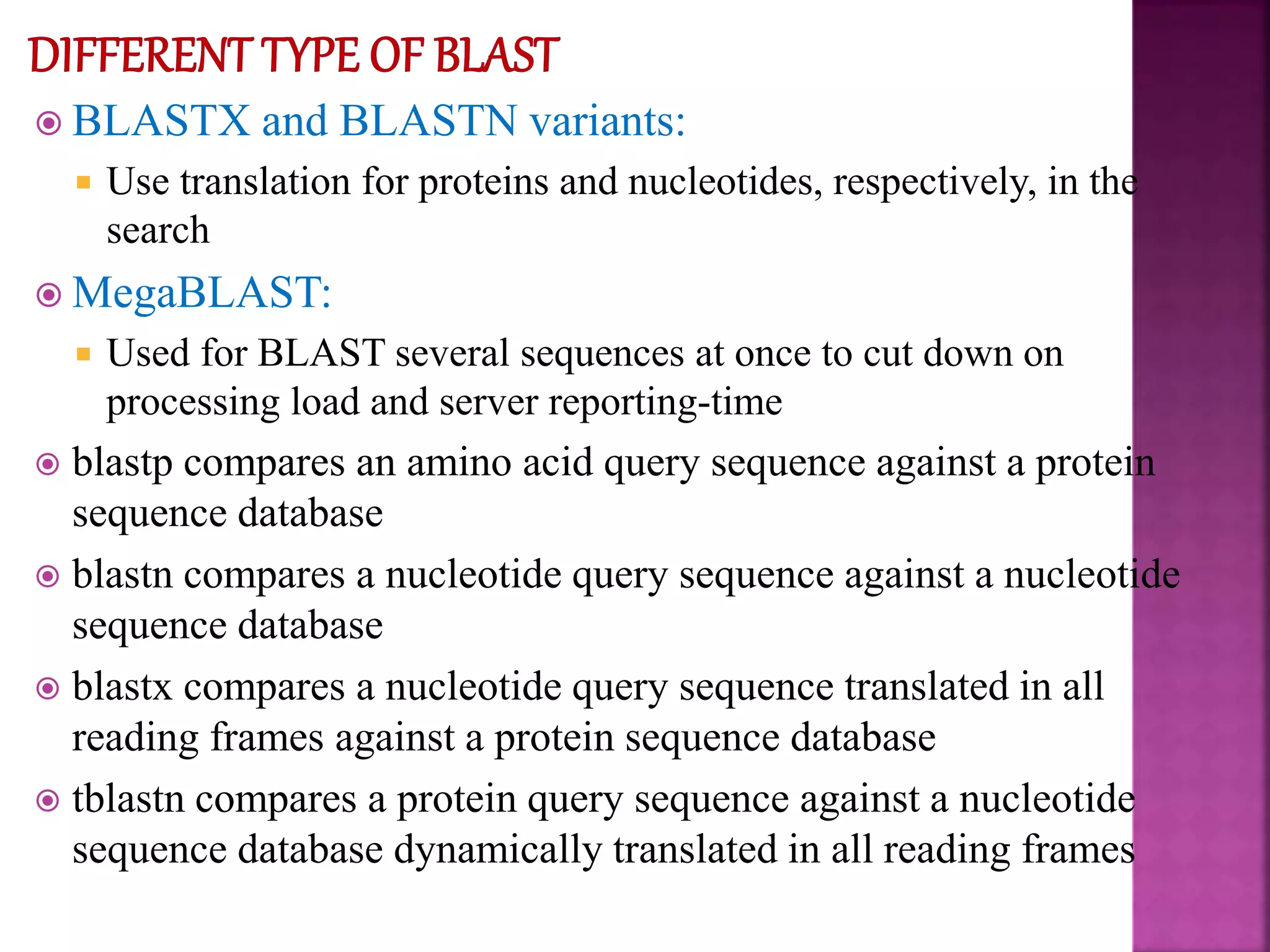
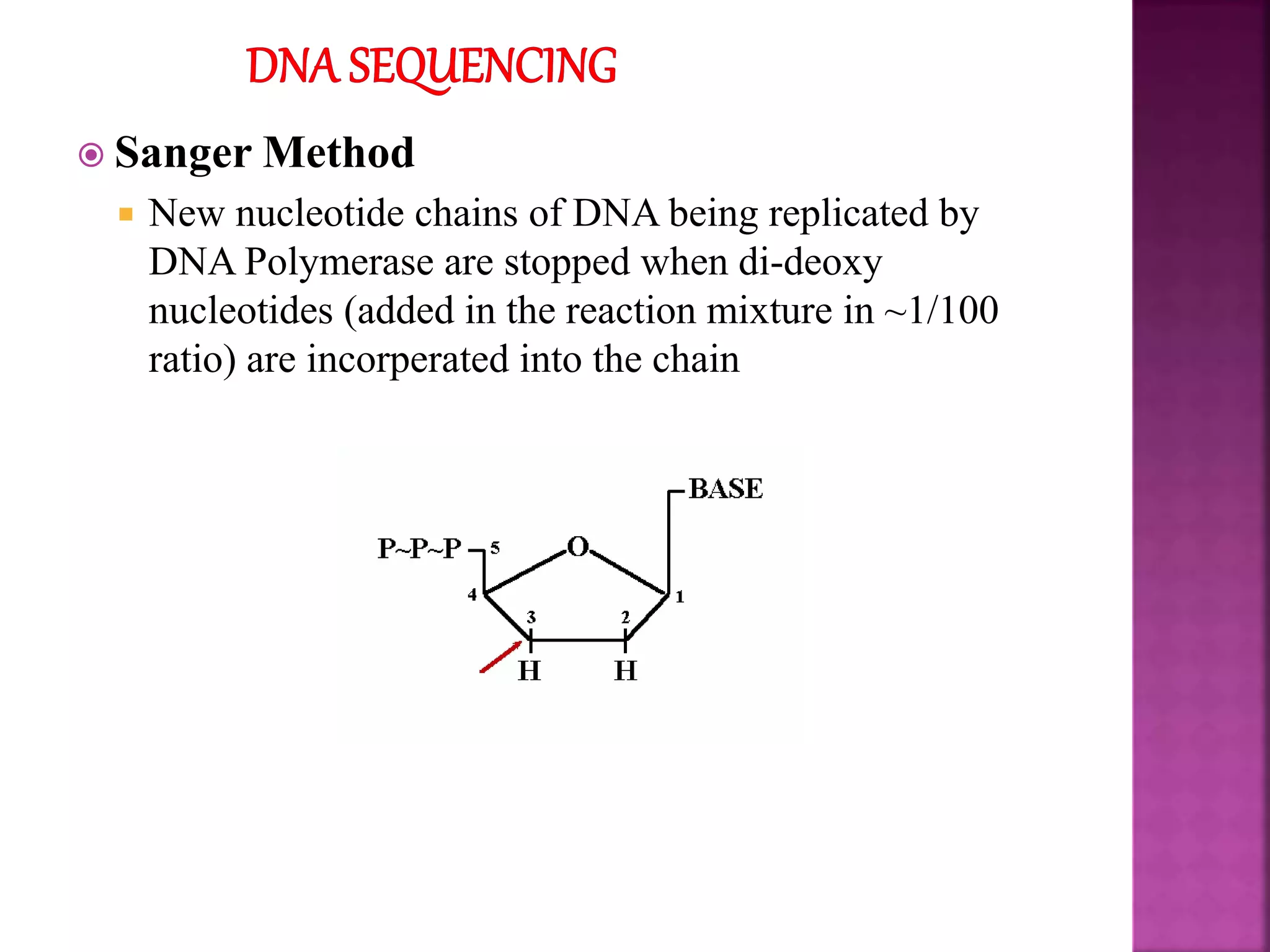
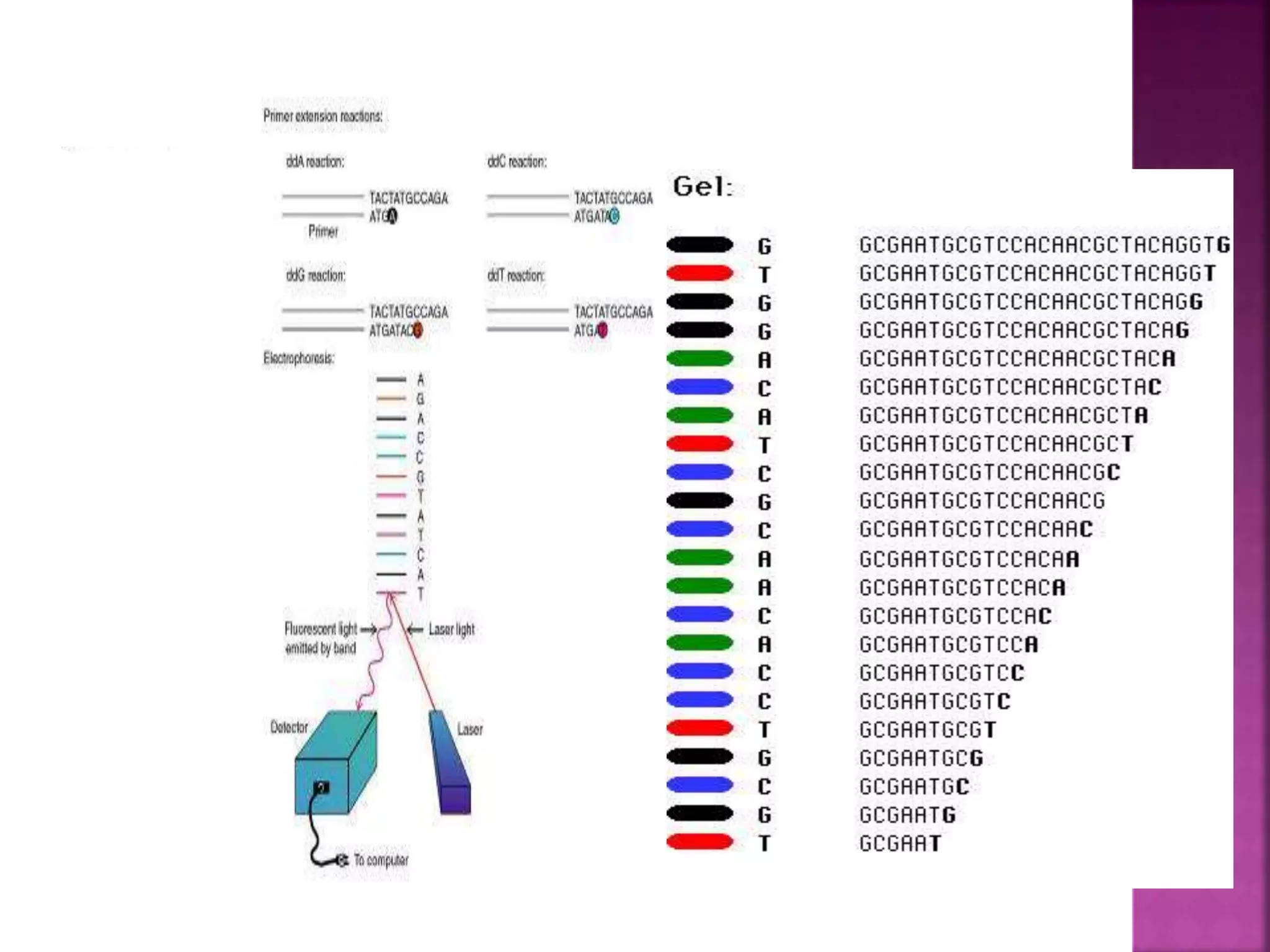
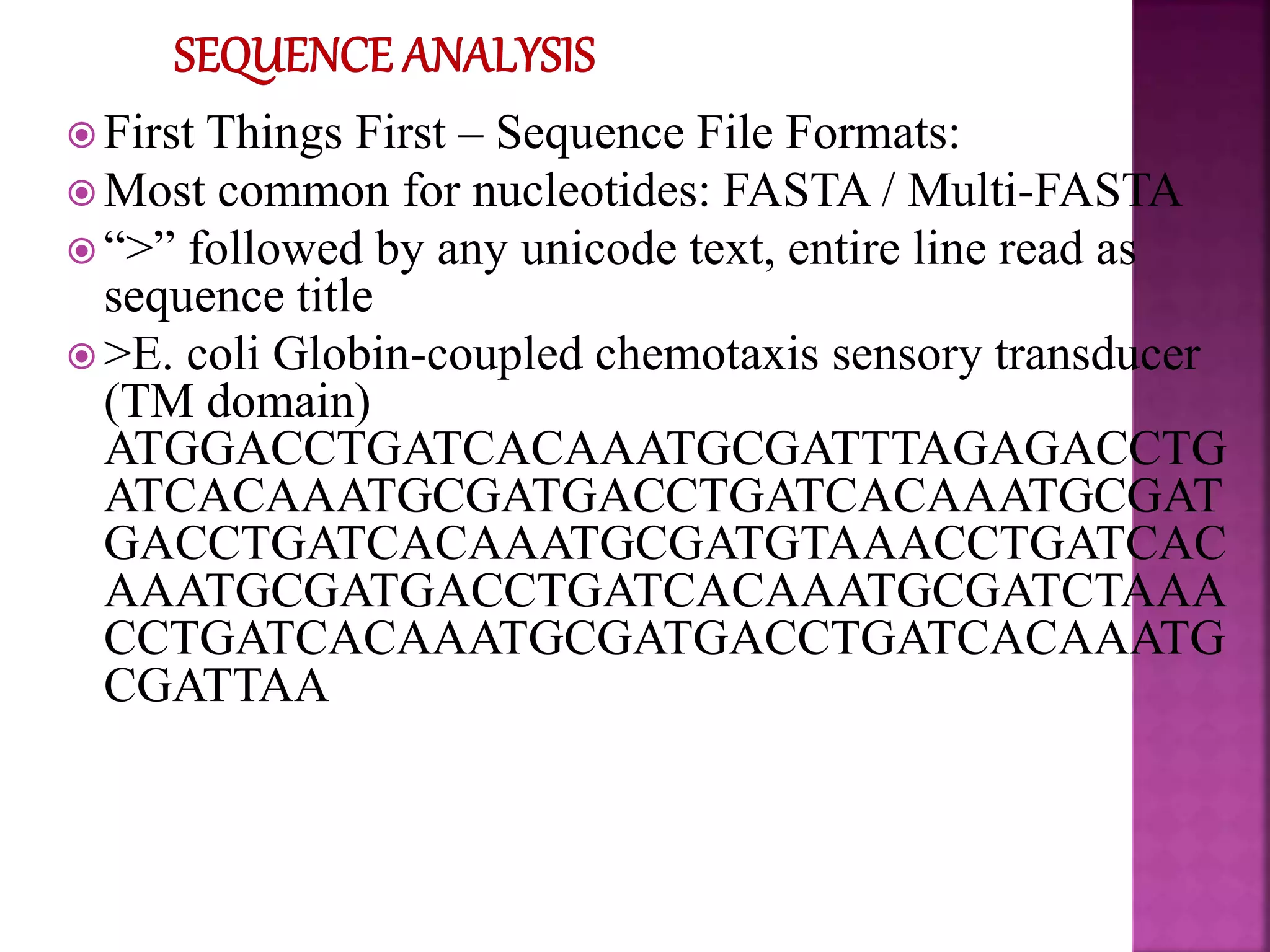
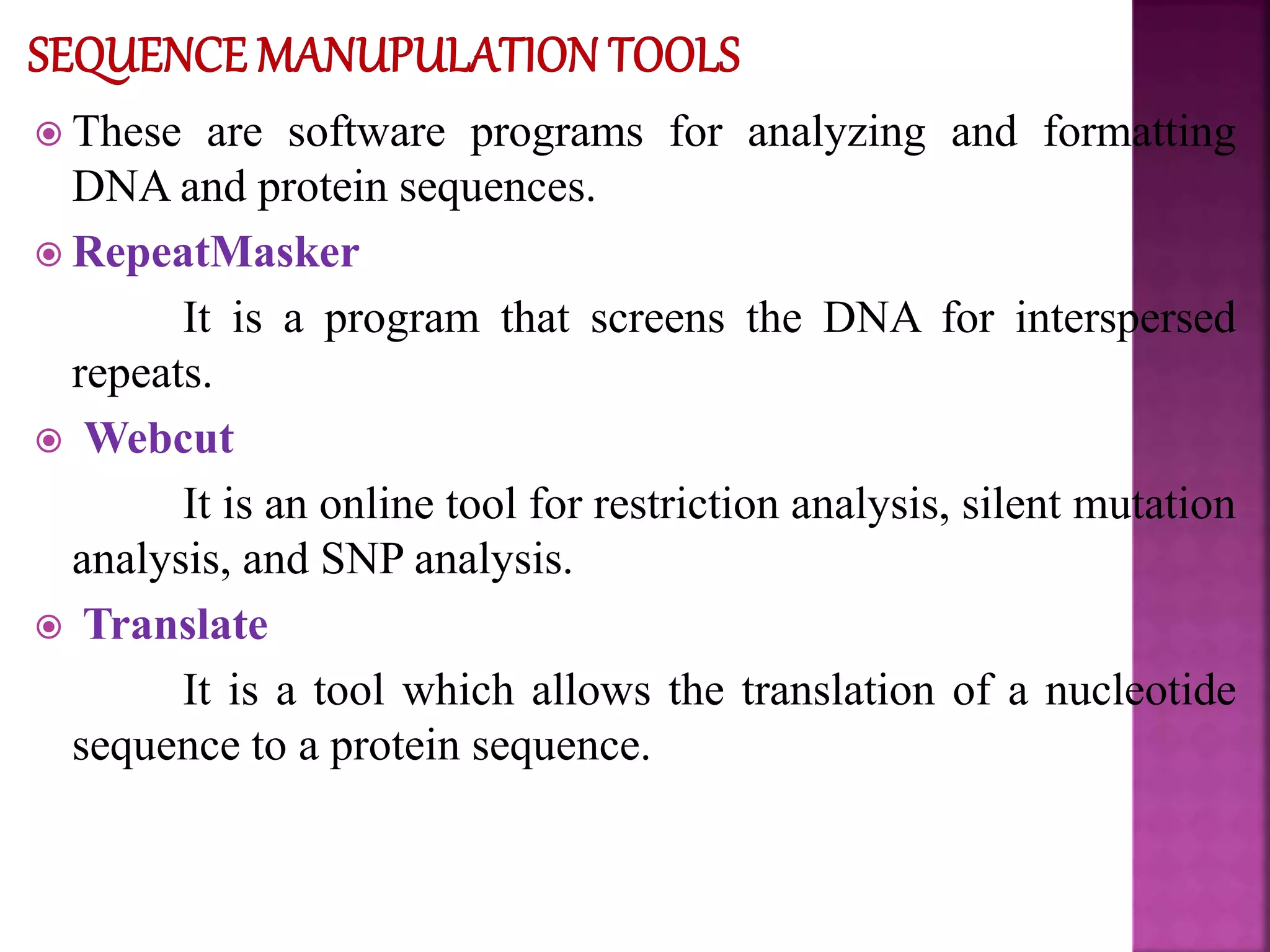
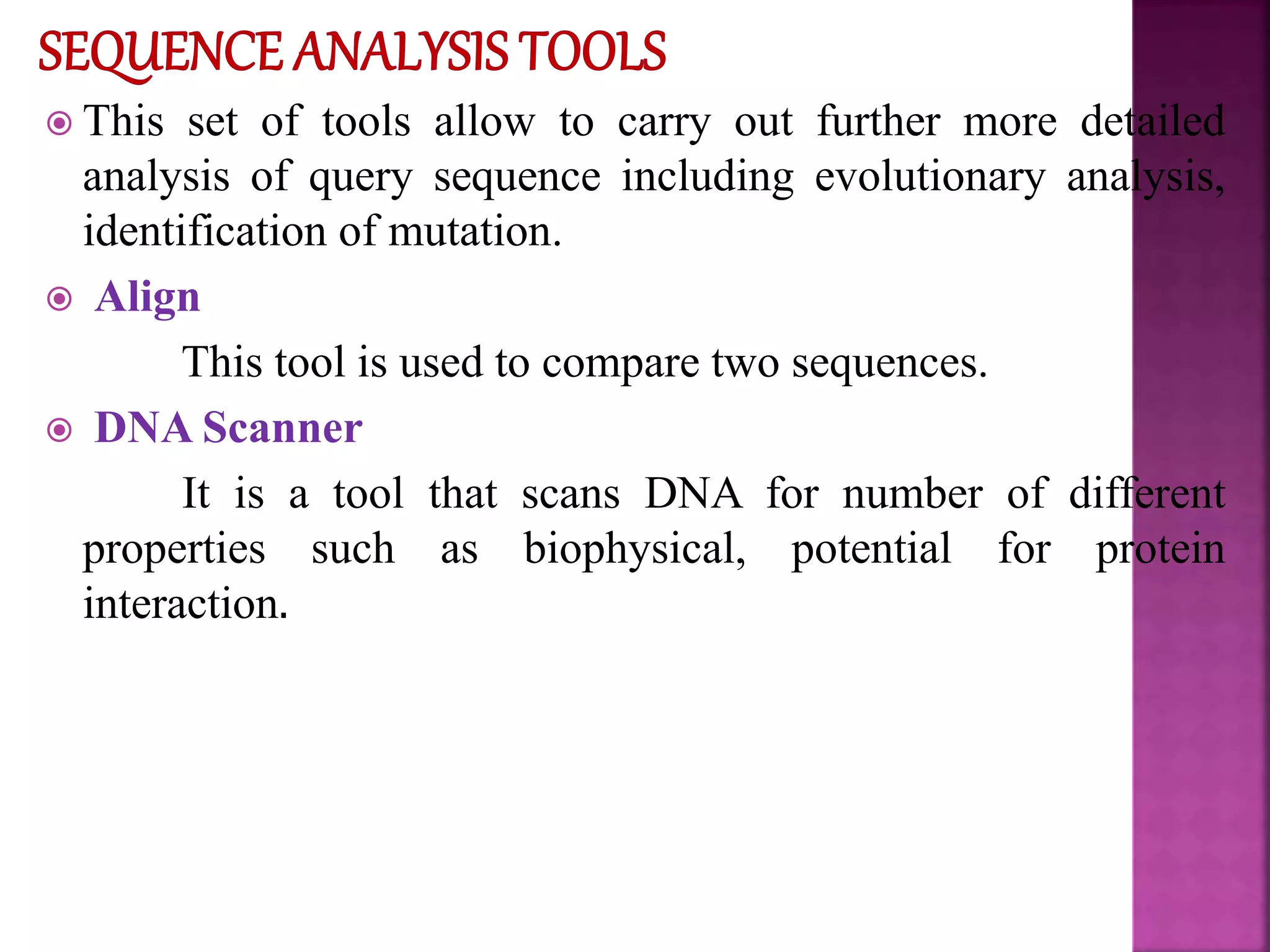
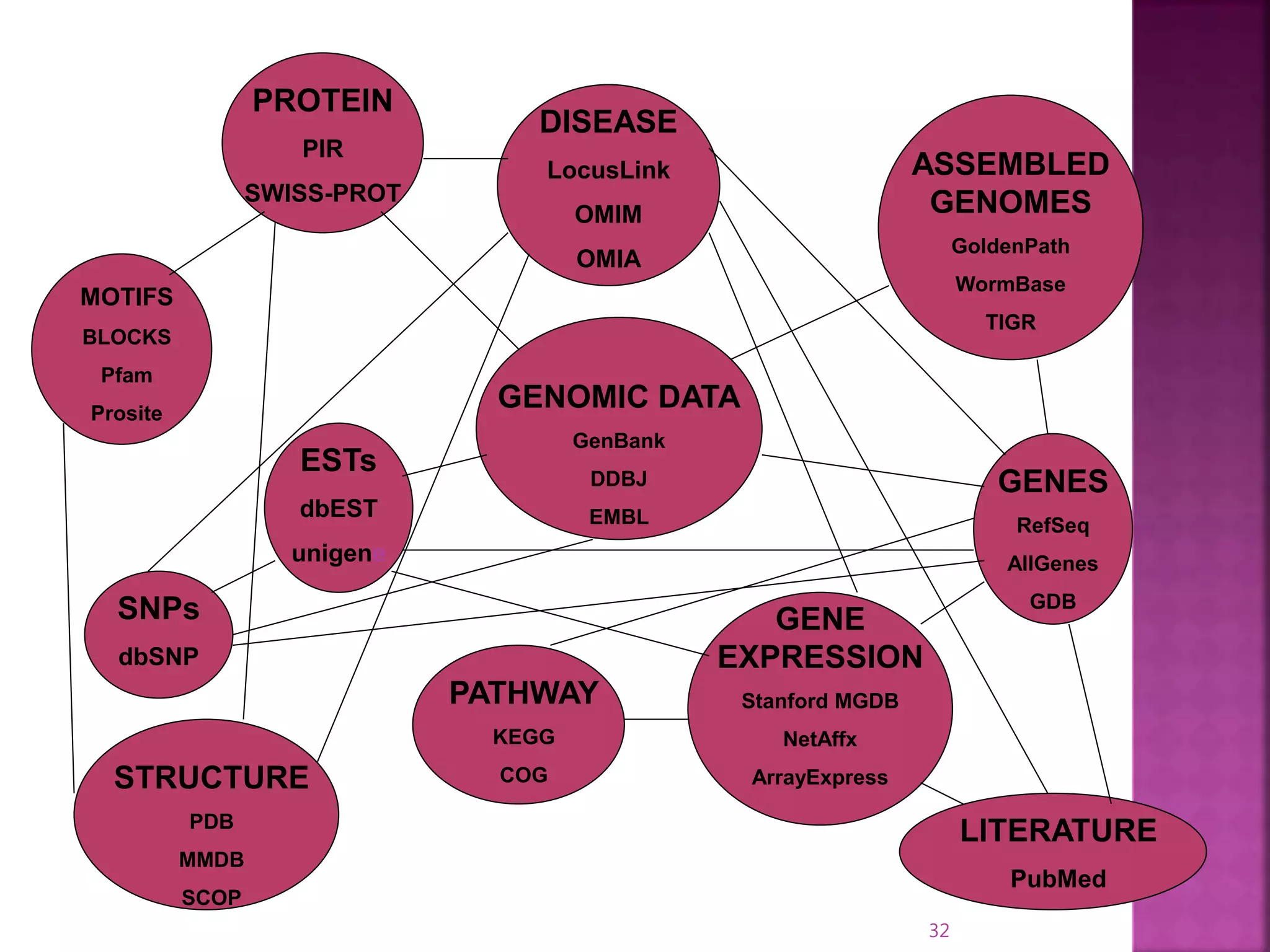
The document discusses bioinformatics tools used for analyzing biological data. It begins with an introduction to bioinformatics and then describes several categories of tools: biological databases for storing genomic and protein data; homology tools for sequence alignment and comparison; protein function analysis tools; structural analysis tools; and sequence manipulation and analysis tools. Common tools discussed include BLAST, FASTA, ClustalW, and databases like GenBank. The document concludes by covering applications of bioinformatics in areas like molecular modeling, medicine, and computation.