prediction methods for ORF
•Download as PPTX, PDF•
21 likes•7,388 views
prediction methods for ORF. homology based method, ab initio based . this slide is fully content with prediction methods for ORF.
Report
Share
Report
Share
Recommended
STRUCTURAL GENOMICS, FUNCTIONAL GENOMICS, COMPARATIVE GENOMICS

The presentation gives you a complete idea about the concepts of structural genomics, functional genomics and comparative genomics
MULTIPLE SEQUENCE ALIGNMENT

Descibes about the patterns in pairwise alignment,multiple sequence alignment and genetic algorithm.
Recommended
STRUCTURAL GENOMICS, FUNCTIONAL GENOMICS, COMPARATIVE GENOMICS

The presentation gives you a complete idea about the concepts of structural genomics, functional genomics and comparative genomics
MULTIPLE SEQUENCE ALIGNMENT

Descibes about the patterns in pairwise alignment,multiple sequence alignment and genetic algorithm.
Microsatellite

Tandem repeats of DNA is useful topic for forensic specialists, clinical lab personnel, medical students.
Sts

STS stands for sequence tagged site which is short DNA sequence, generally between 100 and 500 bp in length, that is easily recognizable and occurs only once in the chromosome or genome being studied.
Genetic and Physical map of Genome

Introduction
History
Genetic mapping
DNA Markers
Physical mapping
Importance
Drawback
Conclusion
References
uses genetic techniques to construct maps showing the positions of genes and other sequence features on a genome.
Genetic techniques include cross-breeding experiments or, in the case of humans, the examination of family histories (pedigrees).
Finding ORF

Open reading frame is part of reading frame that contains no stop codons or region of amino acids coding triple codons.
ORF starts with start codon and ends at stop codon.
Clustal W - Multiple Sequence alignment 

KEGG GenomeNet - Introduction
ClustalW - Introduction
- Algorithm
- Flowchart
Multiple Alignment Method
- Introduction
ClustalW – Work process
Blast and fasta

blast and fasta are two softwares in bioinformatics, blast usually used for similarity checking
Genome annotation 2013

Genome annotation, NGS sequence data, decoding sequence information, The genome contains all the biological information required to build and maintain any given living organism.
Physical mapping

A physical map of a chromosome or a genome that shows the physical locations of genes and other DNA sequences of interest. Physical maps are used to help scientists identify and isolate genes by positional cloning.
According to the ICSM (Intergovernmental Committee on Surveying and Mapping), there are five different types of maps: General Reference, Topographical, Thematic, Navigation Charts and Cadastral Maps and Plans.
Map based cloning 

Map based cloning or Positional Cloning - Application of Genetic mapping and marker assisted studies
Gene prediction methods vijay

Automated sequencing of genomes require automated gene assignment
Includes detection of open reading frames (ORFs)
Identification of the introns and exons
Gene prediction a very difficult problem in pattern recognition
Coding regions generally do not have conserved sequences
Much progress made with prokaryotic gene prediction
Eukaryotic genes more difficult to predict correctly
More Related Content
What's hot
Microsatellite

Tandem repeats of DNA is useful topic for forensic specialists, clinical lab personnel, medical students.
Sts

STS stands for sequence tagged site which is short DNA sequence, generally between 100 and 500 bp in length, that is easily recognizable and occurs only once in the chromosome or genome being studied.
Genetic and Physical map of Genome

Introduction
History
Genetic mapping
DNA Markers
Physical mapping
Importance
Drawback
Conclusion
References
uses genetic techniques to construct maps showing the positions of genes and other sequence features on a genome.
Genetic techniques include cross-breeding experiments or, in the case of humans, the examination of family histories (pedigrees).
Finding ORF

Open reading frame is part of reading frame that contains no stop codons or region of amino acids coding triple codons.
ORF starts with start codon and ends at stop codon.
Clustal W - Multiple Sequence alignment 

KEGG GenomeNet - Introduction
ClustalW - Introduction
- Algorithm
- Flowchart
Multiple Alignment Method
- Introduction
ClustalW – Work process
Blast and fasta

blast and fasta are two softwares in bioinformatics, blast usually used for similarity checking
Genome annotation 2013

Genome annotation, NGS sequence data, decoding sequence information, The genome contains all the biological information required to build and maintain any given living organism.
Physical mapping

A physical map of a chromosome or a genome that shows the physical locations of genes and other DNA sequences of interest. Physical maps are used to help scientists identify and isolate genes by positional cloning.
According to the ICSM (Intergovernmental Committee on Surveying and Mapping), there are five different types of maps: General Reference, Topographical, Thematic, Navigation Charts and Cadastral Maps and Plans.
Map based cloning 

Map based cloning or Positional Cloning - Application of Genetic mapping and marker assisted studies
What's hot (20)
Viewers also liked
Gene prediction methods vijay

Automated sequencing of genomes require automated gene assignment
Includes detection of open reading frames (ORFs)
Identification of the introns and exons
Gene prediction a very difficult problem in pattern recognition
Coding regions generally do not have conserved sequences
Much progress made with prokaryotic gene prediction
Eukaryotic genes more difficult to predict correctly
Free software and bioinformatics

an overview of the free software philosophy as it has been applied on the bioinformatics field
Constraints and Global Optimization for Gene Prediction Overlap Resolution

We apply constraints and global optimization to the problem of restricting overlapping of gene predictions for prokaryotic genomes. We investigate existing heuristic methods and show how they may be expressed using Constraint Handling Rules. Furthermore, we integrate existing methods in a global optimization procedure expressed as probabilistic model in the PRISM language. This approach yields an optimal (highest scoring) subset of predictions that satisfy the constraints. Experimental results indicate accuracy comparable to the heuristic approaches.
Translating Cancer Genomes and Transcriptomes for Precision Oncology 

powerpoint adapted from paper written by Sameek Roychowdhury, MD, PhD1,2,3; Arul M. Chinnaiyan, MD, PhD4,5,6,7
DNA Fingerprinting

A brief presentation on DNA Fingerprinting by Ayushi Ghosh, proudly presented by Biomedicz (www.biomedicz.com).
Viewers also liked (20)
Gene Prediction Using Hidden Markov Model and Recurrent Neural Network

Gene Prediction Using Hidden Markov Model and Recurrent Neural Network
Constraints and Global Optimization for Gene Prediction Overlap Resolution

Constraints and Global Optimization for Gene Prediction Overlap Resolution
Session ii g1 overview genomics and gene expression mmc-good

Session ii g1 overview genomics and gene expression mmc-good
Translating Cancer Genomes and Transcriptomes for Precision Oncology 

Translating Cancer Genomes and Transcriptomes for Precision Oncology
Similar to prediction methods for ORF
Genome annotation

After sequencing of the genome has been done, the first thing that comes to mind is "Where are the genes?". Genome annotation is the process of attaching information to the biological sequences. It is an active area of research and it would help scientists a lot to undergo with their wet lab projects once they know the coding parts of a genome.
Gene identification using bioinformatic tools.pptx

Being able to identify genes, compare them, analyze them could be applied in various research areas from medical to industrial.
This ppt is designed for Health science and computational biology students to enable you understand the above mentioned topic.
Apollo Introduction for i5K Groups 2015-10-07

This is an introduction to conducting manual annotation efforts using Apollo. This webinar was offered to members of the i5K Research community on 2015-10-07.
Introduction to Apollo: i5K E affinis

Apollo is a web-based application that supports and enables collaborative genome curation in real time, allowing teams of curators to improve on existing automated gene models through an intuitive interface. Apollo allows researchers to break down large amounts of data into manageable portions to mobilize groups of researchers with shared interests.
The i5K, an initiative to sequence the genomes of 5,000 insect and related arthropod species, is a broad and inclusive effort that seeks to involve scientists from around the world in their genome curation process, and Apollo is serving as the platform to empower this community.
This presentation is an introduction to Apollo for the members of the i5K Pilot Project on Eurytemora affinis
Finding genes

ORF is Open Reading Frame that are the Exon sequences which are to be translated in the process of translation
gene prediction programs

BIOINFORMATICS- gene prediction programs
grail, augustus, genscan, hmmgene, mzef
with features, alogrithm, input, output, and species
Introduction to Apollo: A webinar for the i5K Research Community

Apollo is a web-based application that supports and enables collaborative genome curation in real time, allowing teams of curators to improve on existing automated gene models through an intuitive interface. Apollo allows researchers to break down large amounts of data into manageable portions to mobilize groups of researchers with shared interests.
The i5K, an initiative to sequence the genomes of 5,000 insect and related arthropod species, is a broad and inclusive effort that seeks to involve scientists from around the world in their genome curation process, and Apollo is serving as the platform to empower this community.
This presentation is an introduction to Apollo for the members of the i5K Pilot Project Species.
Similar to prediction methods for ORF (20)
BTC 506 Gene Identification using Bioinformatic Tools-230302130331.pptx

BTC 506 Gene Identification using Bioinformatic Tools-230302130331.pptx
Gene identification using bioinformatic tools.pptx

Gene identification using bioinformatic tools.pptx
Introduction to Apollo: A webinar for the i5K Research Community

Introduction to Apollo: A webinar for the i5K Research Community
Recently uploaded
Salas, V. (2024) "John of St. Thomas (Poinsot) on the Science of Sacred Theol...

I Introduction
II Subalternation and Theology
III Theology and Dogmatic Declarations
IV The Mixed Principles of Theology
V Virtual Revelation: The Unity of Theology
VI Theology as a Natural Science
VII Theology’s Certitude
VIII Conclusion
Notes
Bibliography
All the contents are fully attributable to the author, Doctor Victor Salas. Should you wish to get this text republished, get in touch with the author or the editorial committee of the Studia Poinsotiana. Insofar as possible, we will be happy to broker your contact.
What is greenhouse gasses and how many gasses are there to affect the Earth.

What are greenhouse gasses how they affect the earth and its environment what is the future of the environment and earth how the weather and the climate effects.
DERIVATION OF MODIFIED BERNOULLI EQUATION WITH VISCOUS EFFECTS AND TERMINAL V...

In this book, we use conservation of energy techniques on a fluid element to derive the Modified Bernoulli equation of flow with viscous or friction effects. We derive the general equation of flow/ velocity and then from this we derive the Pouiselle flow equation, the transition flow equation and the turbulent flow equation. In the situations where there are no viscous effects , the equation reduces to the Bernoulli equation. From experimental results, we are able to include other terms in the Bernoulli equation. We also look at cases where pressure gradients exist. We use the Modified Bernoulli equation to derive equations of flow rate for pipes of different cross sectional areas connected together. We also extend our techniques of energy conservation to a sphere falling in a viscous medium under the effect of gravity. We demonstrate Stokes equation of terminal velocity and turbulent flow equation. We look at a way of calculating the time taken for a body to fall in a viscous medium. We also look at the general equation of terminal velocity.
Seminar of U.V. Spectroscopy by SAMIR PANDA

Spectroscopy is a branch of science dealing the study of interaction of electromagnetic radiation with matter.
Ultraviolet-visible spectroscopy refers to absorption spectroscopy or reflect spectroscopy in the UV-VIS spectral region.
Ultraviolet-visible spectroscopy is an analytical method that can measure the amount of light received by the analyte.
Orion Air Quality Monitoring Systems - CWS

Professional air quality monitoring systems provide immediate, on-site data for analysis, compliance, and decision-making.
Monitor common gases, weather parameters, particulates.
bordetella pertussis.................................ppt

Bordettela is a gram negative cocobacilli spread by air born drop let
(May 29th, 2024) Advancements in Intravital Microscopy- Insights for Preclini...

(May 29th, 2024) Advancements in Intravital Microscopy- Insights for Preclini...Scintica Instrumentation
Intravital microscopy (IVM) is a powerful tool utilized to study cellular behavior over time and space in vivo. Much of our understanding of cell biology has been accomplished using various in vitro and ex vivo methods; however, these studies do not necessarily reflect the natural dynamics of biological processes. Unlike traditional cell culture or fixed tissue imaging, IVM allows for the ultra-fast high-resolution imaging of cellular processes over time and space and were studied in its natural environment. Real-time visualization of biological processes in the context of an intact organism helps maintain physiological relevance and provide insights into the progression of disease, response to treatments or developmental processes.
In this webinar we give an overview of advanced applications of the IVM system in preclinical research. IVIM technology is a provider of all-in-one intravital microscopy systems and solutions optimized for in vivo imaging of live animal models at sub-micron resolution. The system’s unique features and user-friendly software enables researchers to probe fast dynamic biological processes such as immune cell tracking, cell-cell interaction as well as vascularization and tumor metastasis with exceptional detail. This webinar will also give an overview of IVM being utilized in drug development, offering a view into the intricate interaction between drugs/nanoparticles and tissues in vivo and allows for the evaluation of therapeutic intervention in a variety of tissues and organs. This interdisciplinary collaboration continues to drive the advancements of novel therapeutic strategies.
Nucleic Acid-its structural and functional complexity.

This presentation explores a brief idea about the structural and functional attributes of nucleotides, the structure and function of genetic materials along with the impact of UV rays and pH upon them.
Recently uploaded (20)
erythropoiesis-I_mechanism& clinical significance.pptx

erythropoiesis-I_mechanism& clinical significance.pptx
Salas, V. (2024) "John of St. Thomas (Poinsot) on the Science of Sacred Theol...

Salas, V. (2024) "John of St. Thomas (Poinsot) on the Science of Sacred Theol...
What is greenhouse gasses and how many gasses are there to affect the Earth.

What is greenhouse gasses and how many gasses are there to affect the Earth.
DERIVATION OF MODIFIED BERNOULLI EQUATION WITH VISCOUS EFFECTS AND TERMINAL V...

DERIVATION OF MODIFIED BERNOULLI EQUATION WITH VISCOUS EFFECTS AND TERMINAL V...
bordetella pertussis.................................ppt

bordetella pertussis.................................ppt
Lateral Ventricles.pdf very easy good diagrams comprehensive

Lateral Ventricles.pdf very easy good diagrams comprehensive
(May 29th, 2024) Advancements in Intravital Microscopy- Insights for Preclini...

(May 29th, 2024) Advancements in Intravital Microscopy- Insights for Preclini...
Nucleic Acid-its structural and functional complexity.

Nucleic Acid-its structural and functional complexity.
prediction methods for ORF
- 1. BY:- BY:- KARAMVEER M.Sc. LIFE SCIENCES WITH SPECIALISATION BIOINFORMATICS (2015-17) WEL-COME
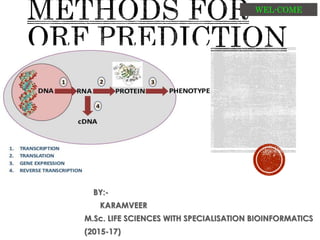
- 2. From a genomic DNA sequence we want to predict the regions that will encode for a protein: the genes. • Gene finding is about detecting these coding regions and infer the gene structure starting from genomic DNA sequences. an open reading frame (ORF) is the part of a reading frame that has the potential to code for a protein or peptide. An ORF is a continuous stretch of codons that do not contain a stop codon • We need to distinguish coding from non-coding regions using properties specific to each type of DNA region. • Gene finding is not an easy task! • DNA sequence signals have low information content. • It is difficult to discriminate real signals from noise (degenerated and highly unspecific signals);
- 4. Simple 1st step in gene findings. Translate genomic sequence in six frames. Identify stop codon in each frame. Regions without stop codons are called “open reading frames” or ORFs. Locate and tag all of the likely ORFs in a sequence. The longest ORF from a methionine codon is a good prediction of a protein encoding sequence.
- 5. The ORF finder is a graphical analysis tool which finds all open reading of a selectable minimum size in a user’s sequence or in a sequence already in the database. This tool identifies all open reading frames using the standard genetic codes. The deduced amino acid sequence can be saved in various format and searched against the sequence database using the blast server. The orf finder should be helpful in preparing complete and accurate sequence.
- 8. Based on sequence similarity of query sequence with annotated genes present in databases. Given a database of sequences of other organism. Search for query sequence in this database . Identify database sequence (known genes) that resemble the query sequence. If the identified sequences are genes , the query sequence is probably (putatively) a gene.
- 9. Basic local alignment search tool. Well known search tool in this category. Strengths:- able to identify biologically relevant genes. Accuracy weakness:- Could not identify genes that code for protein , not present in database. Only 50% genes can be found by homology to other known genes or proteins.
- 10. Uses HMMs to compare DNA sequences to protein sequences at the level of its conceptual translation, regardless of sequencing errors and introns. • Principle: • The exon model used in genewise is a HMM with 3 base states (match, insert, delete) with the addition of more transitions between states to consider frame- shifts. • Intron states have been added to the base model. • Genewise directly compare HMM-profiles of proteins or domains to the gene structure HMM model. • Genewise is a powerful tool, but time consuming. • Requires strong similarities (>70% identity) to produce good predictions. • Genewise is part of the Wise2 package:
- 11. Computational prediction that use most elementary information. Can predict both eukaryotic and prokaryotic genes. Predict genes based on the given sequence alone. It works on two major features associated with genes:- 1. Gene signals 2. Gene content
- 14. • Hidden Markov Models (HMMs):- • HMMs use a probabilistic framework to infer the probability that a sequence correspond to a real signal. • Neural Networks (NNs): • NNs are trained with positive and negative examples. NNs ”discover” the features that distinguish the two sets. • . The gene structure information is separated into several classes of features such as hexamer frequencies, splice sites, and GC composition. Example: NN for acceptor sites, the perceptron, (Horton and Kanehisa, 1992)
- 15. Neural network recognizing coding potential • Incorporates genomic context information (splice junctions, start and stop codons , poly-A signals) • Not appropriate for sequences without genomic context • http://compbio.ornl.gov • Human, Mouse, Drosophila, Arabidopsis, and E. coli
- 16. accuracy of a prediction program can be evaluated using parameters such as sensitivity and specificity. To describe the concept of sensitivity and specificity accurately, four features are used:- true positive (TP), which is a correctly predicted feature; false positive (FP), which is an incorrectly predicted feature; false negative (FN), which is a missed feature; and true negative (TN), which is the correctly predicted absence of a feature.
- 19. Conclusion
