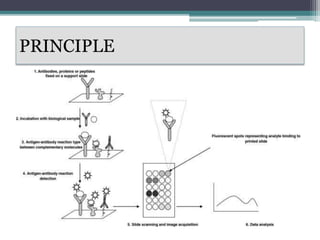
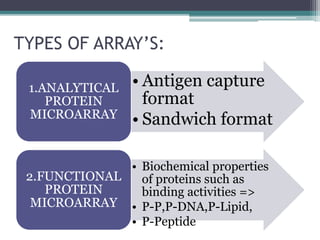
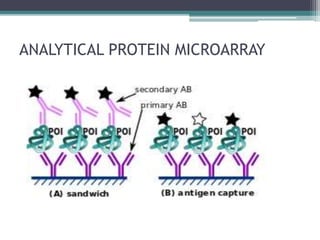
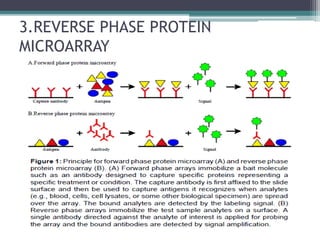
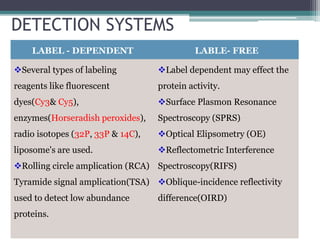
Protein microarrays allow high-throughput analysis of protein interactions and functions. They consist of large numbers of capture proteins immobilized on a surface to which labeled probe molecules are added to detect reactions by fluorescence. There are analytical arrays to study protein binding properties and functional arrays containing full-length proteins to assay enzymatic activity and detect antibodies. Protein microarrays have applications in diagnostics, proteomics, analyzing protein interactions and functions, antibody characterization, and treatment development.