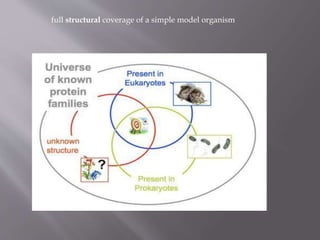
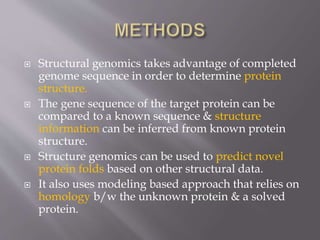
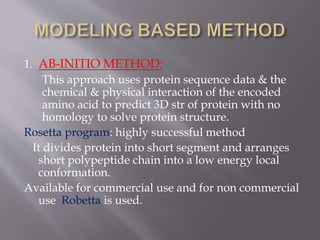
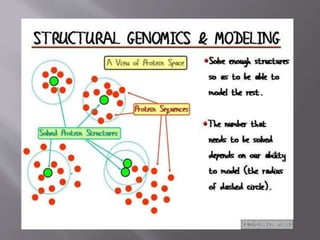
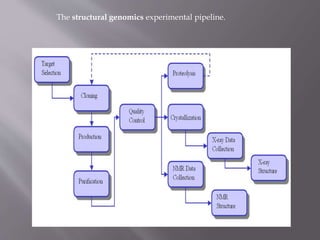
Structural genomics aims to determine the 3D structure of all proteins in a genome. It uses high-throughput methods like X-ray crystallography and NMR on a genomic scale. This allows determination of protein structures for entire proteomes. It provides insights into protein function and can aid drug discovery by identifying potential drug targets like in Mycobacterium tuberculosis. Structural genomics leverages completed genome sequences to clone and express all encoded proteins for structural characterization.