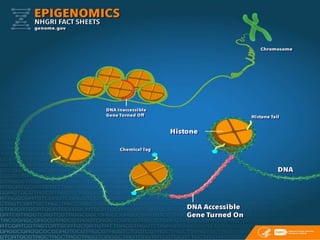
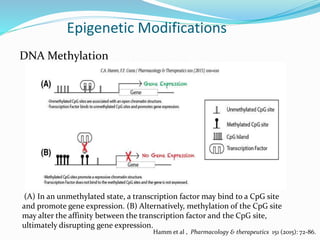
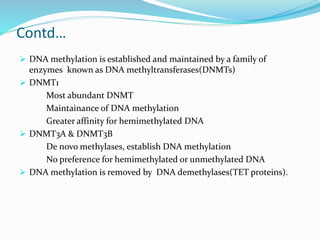
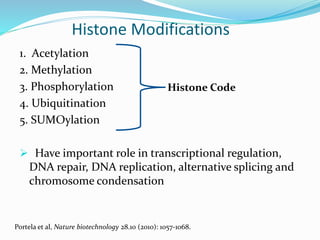
This document discusses epigenetics and epigenetic modifications. It begins by defining the epigenome and how it can change without altering DNA sequence. The main epigenetic modifications discussed are DNA methylation and various histone modifications such as acetylation, methylation, phosphorylation, ubiquitination, and SUMOylation. These modifications can impact gene expression and transcription. The document then examines how epigenetic abnormalities contribute to various diseases like cancer, diabetes, and neurological disorders. Finally, it reviews some FDA-approved epigenetic therapies that target DNA methyltransferases and histone deacetylases, as well as epigenetic drugs currently in clinical trials.