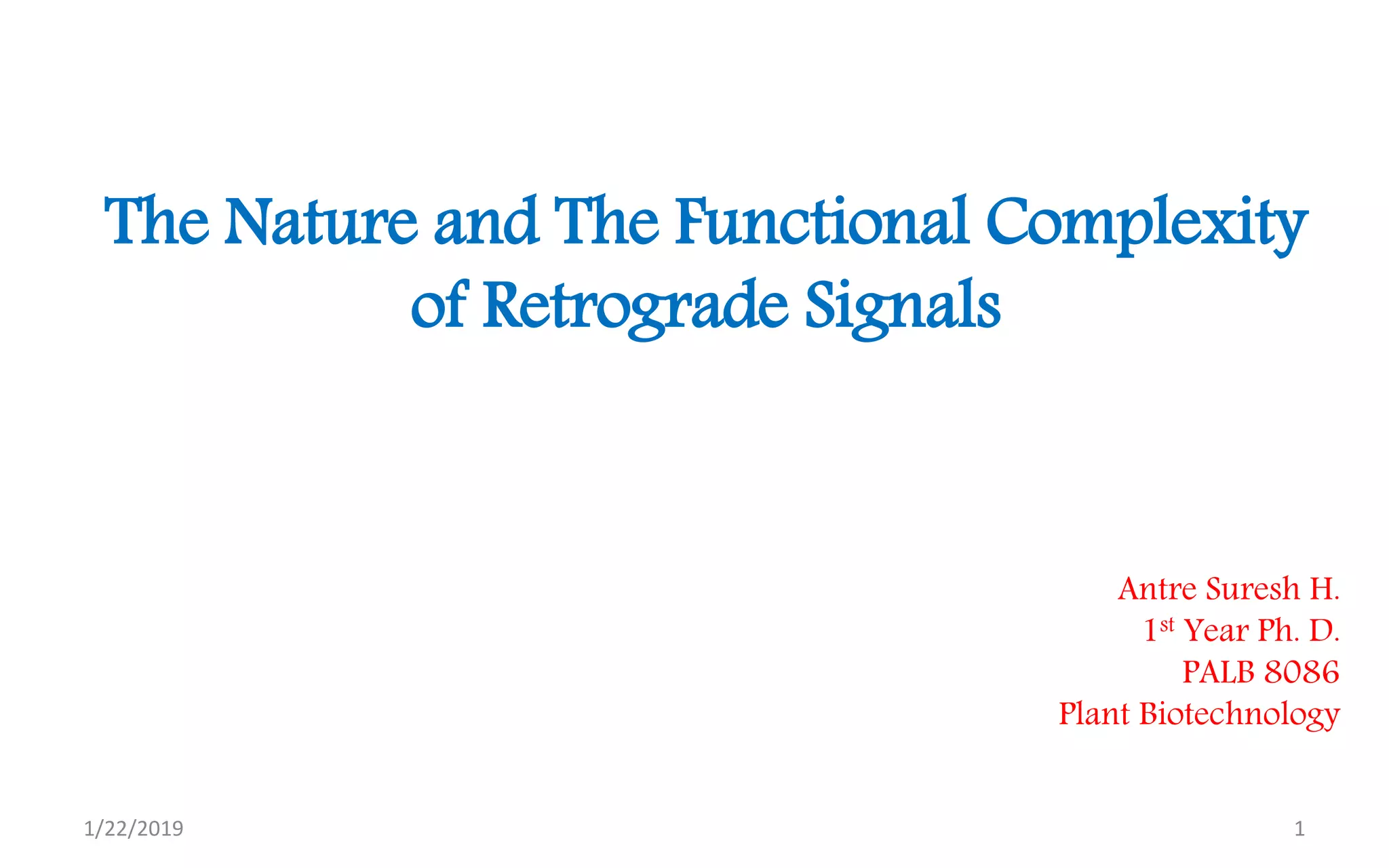
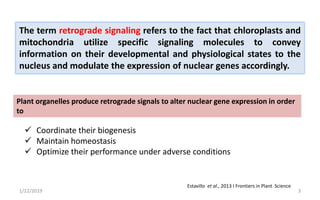
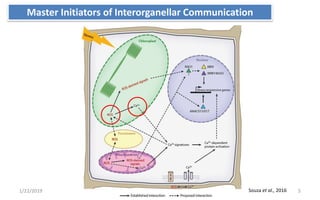
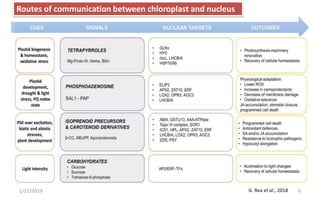
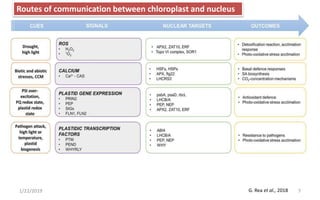
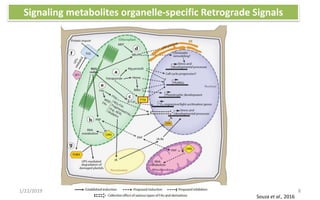
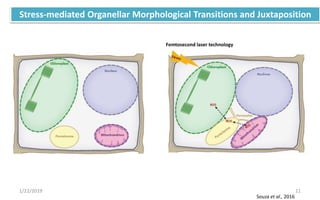
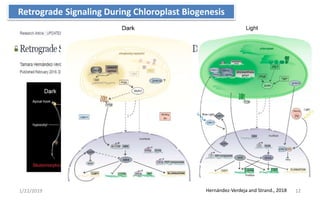
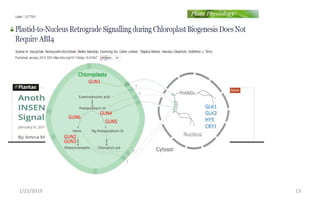
The document discusses retrograde signaling in plants, highlighting its role in coordinating nuclear gene expression based on the developmental and physiological states of chloroplasts and mitochondria. It identifies two primary networks of signals — biogenic and operational — that communicate environmental responses and developmental cues to the nucleus. Additionally, it outlines the complexity of interorganellar communication and the significance of signaling metabolites and master initiators like Ca2+ and ROS in this process.