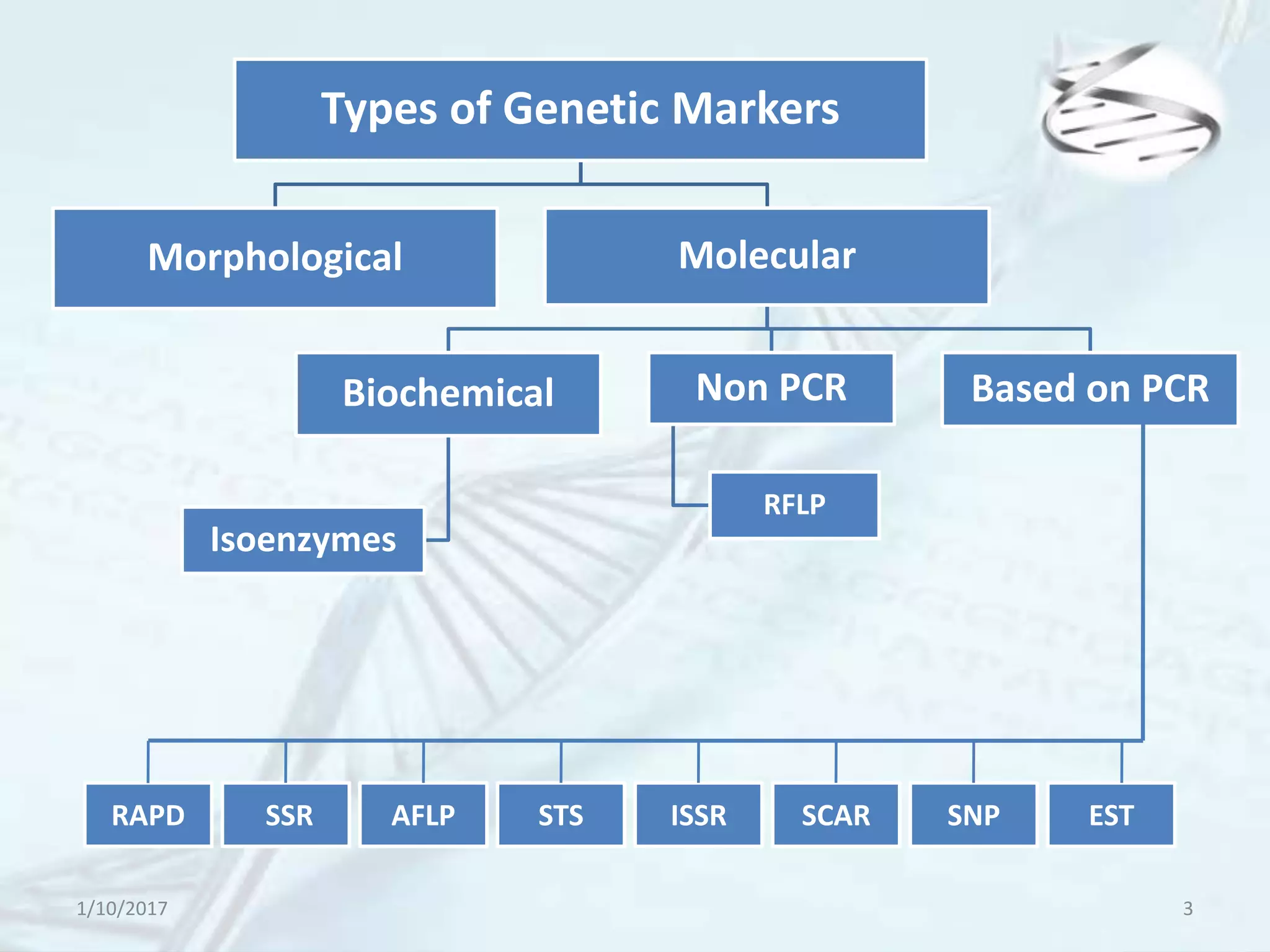
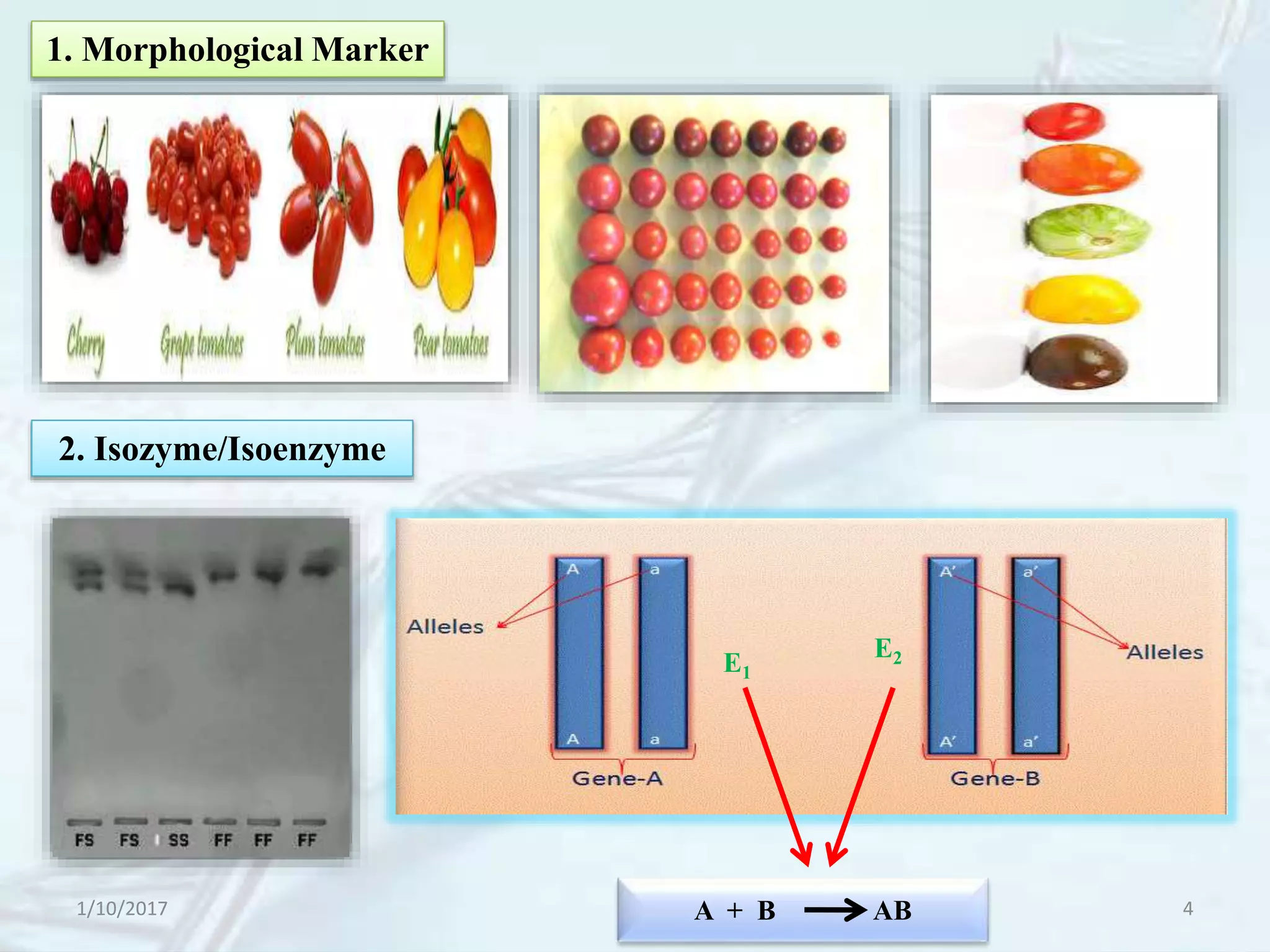
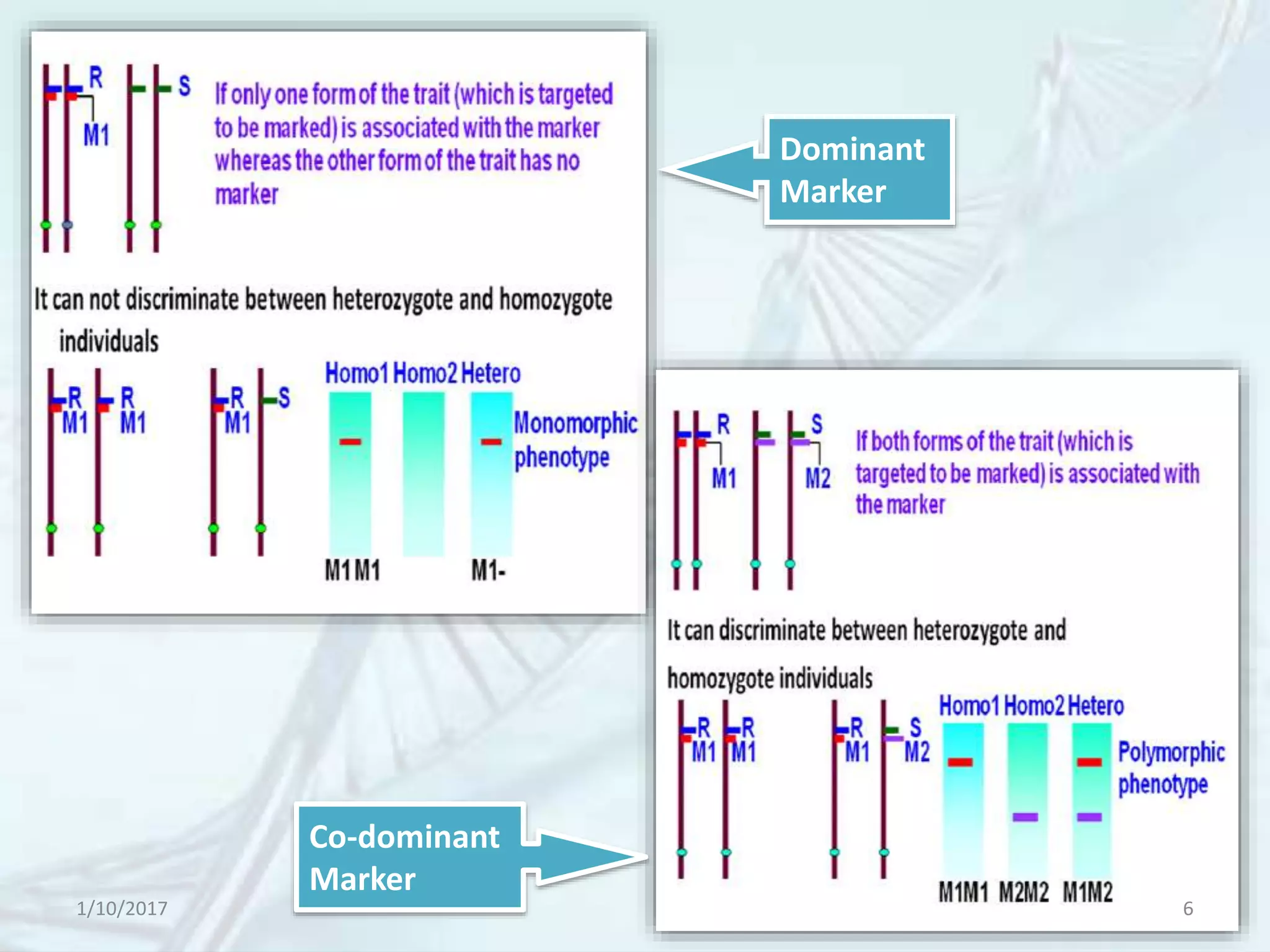
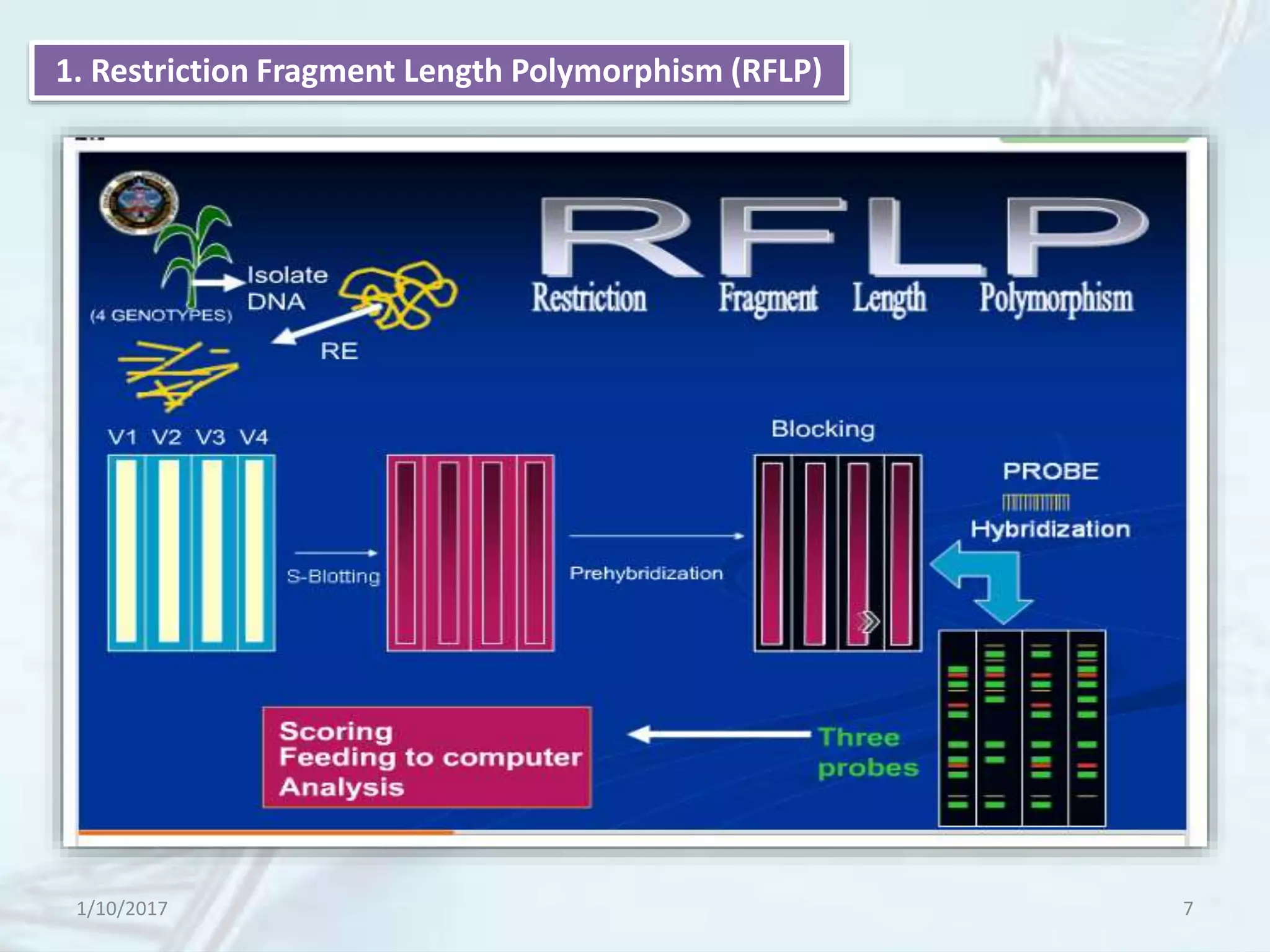
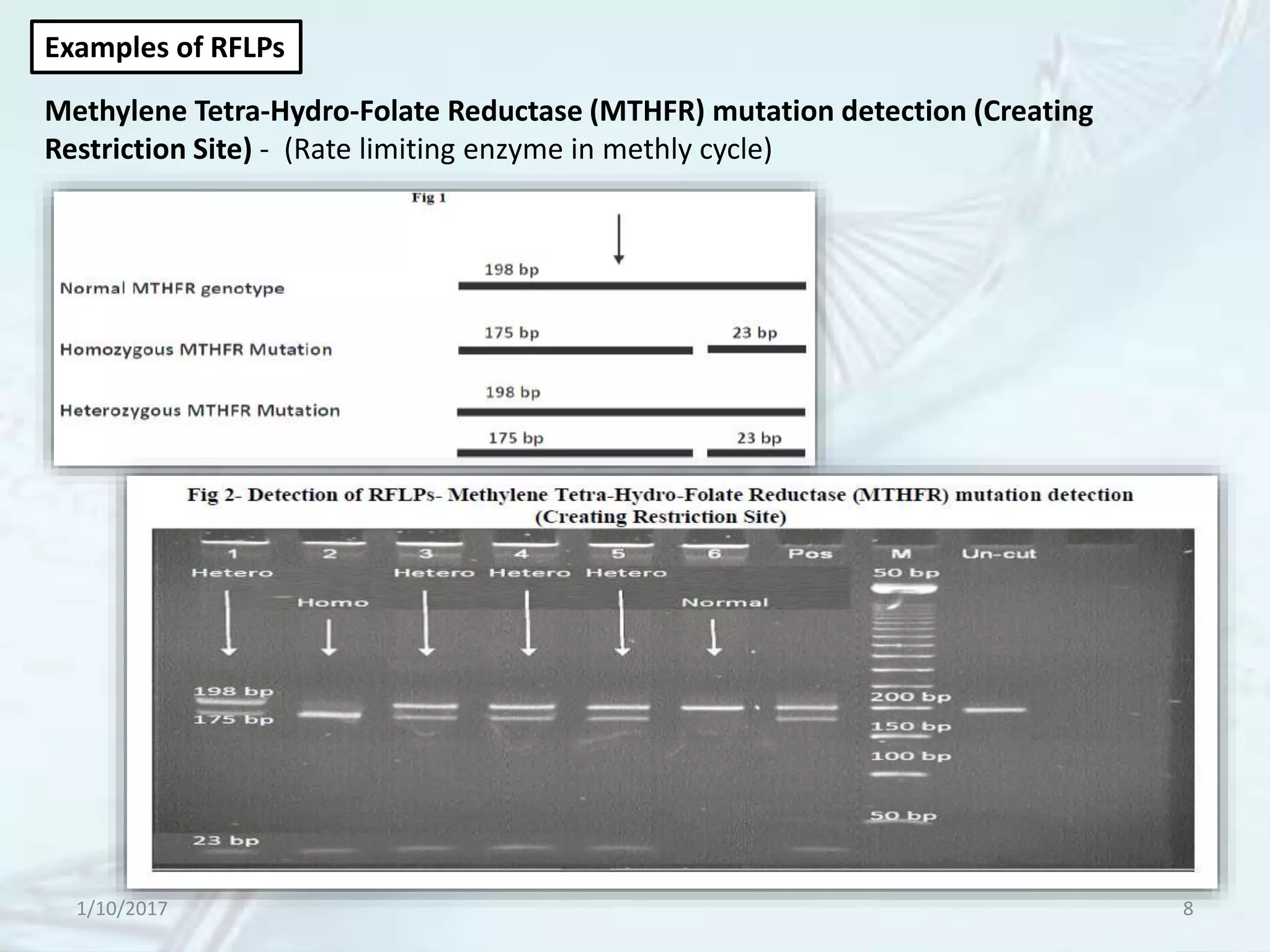
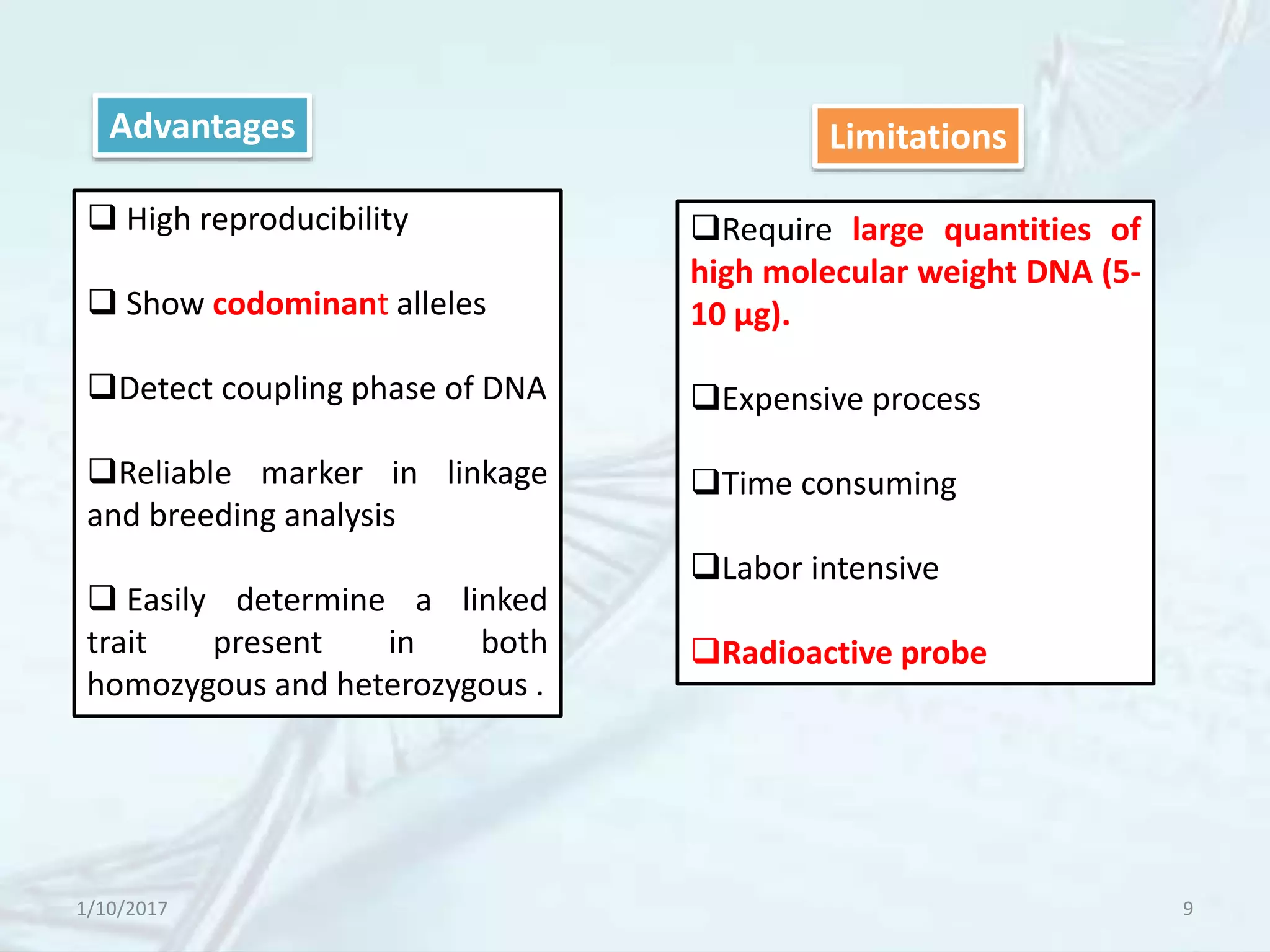
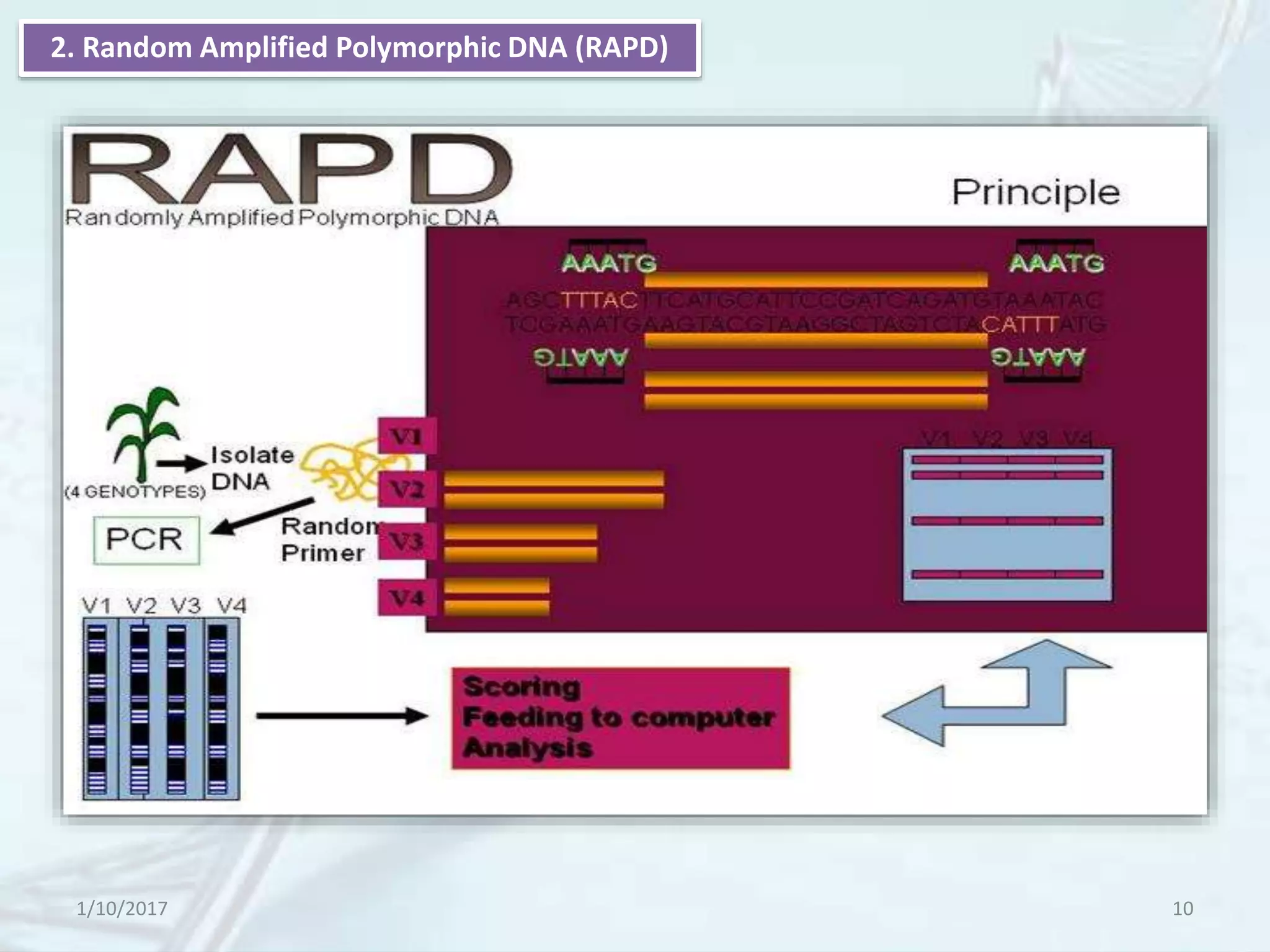
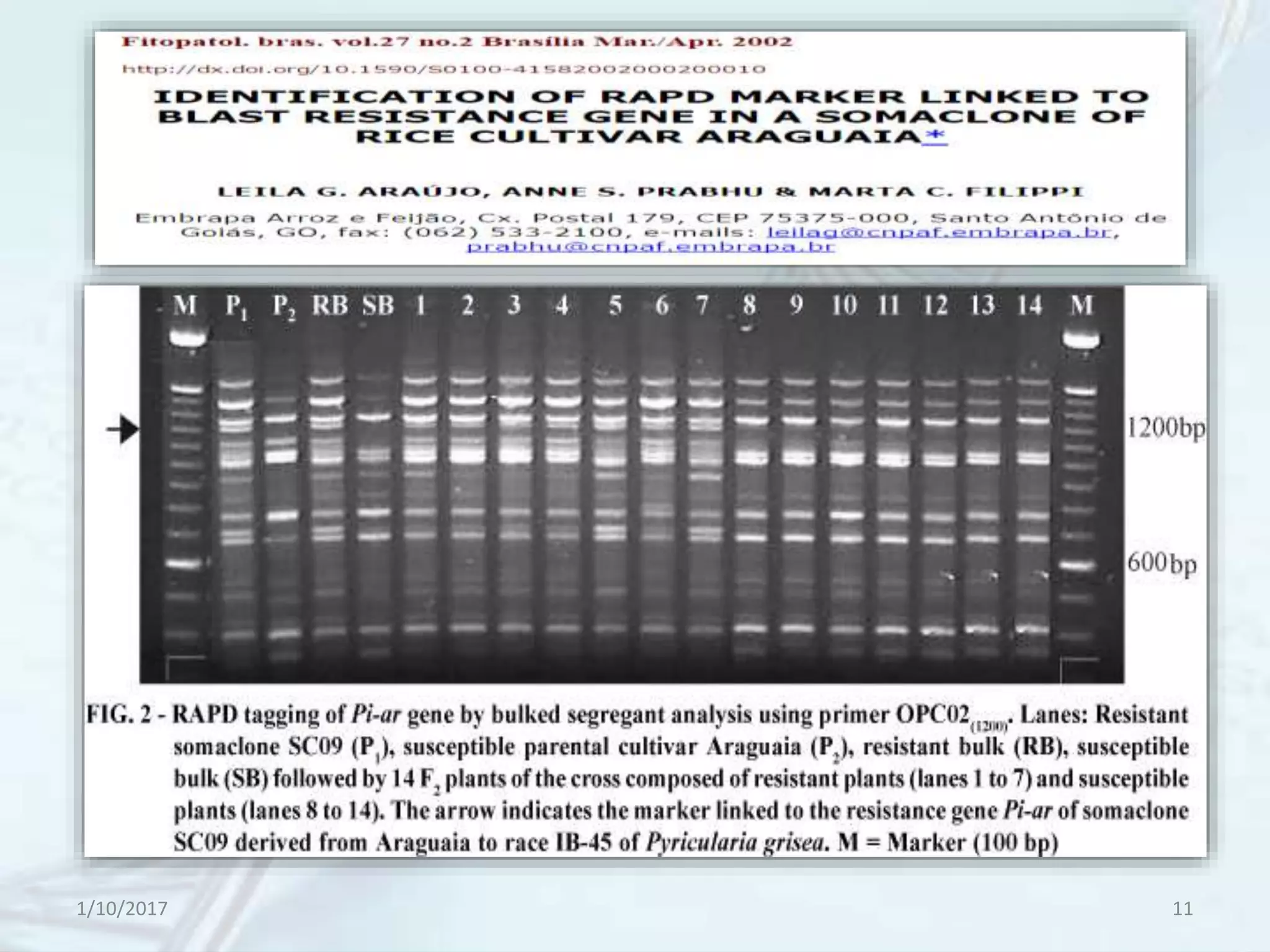
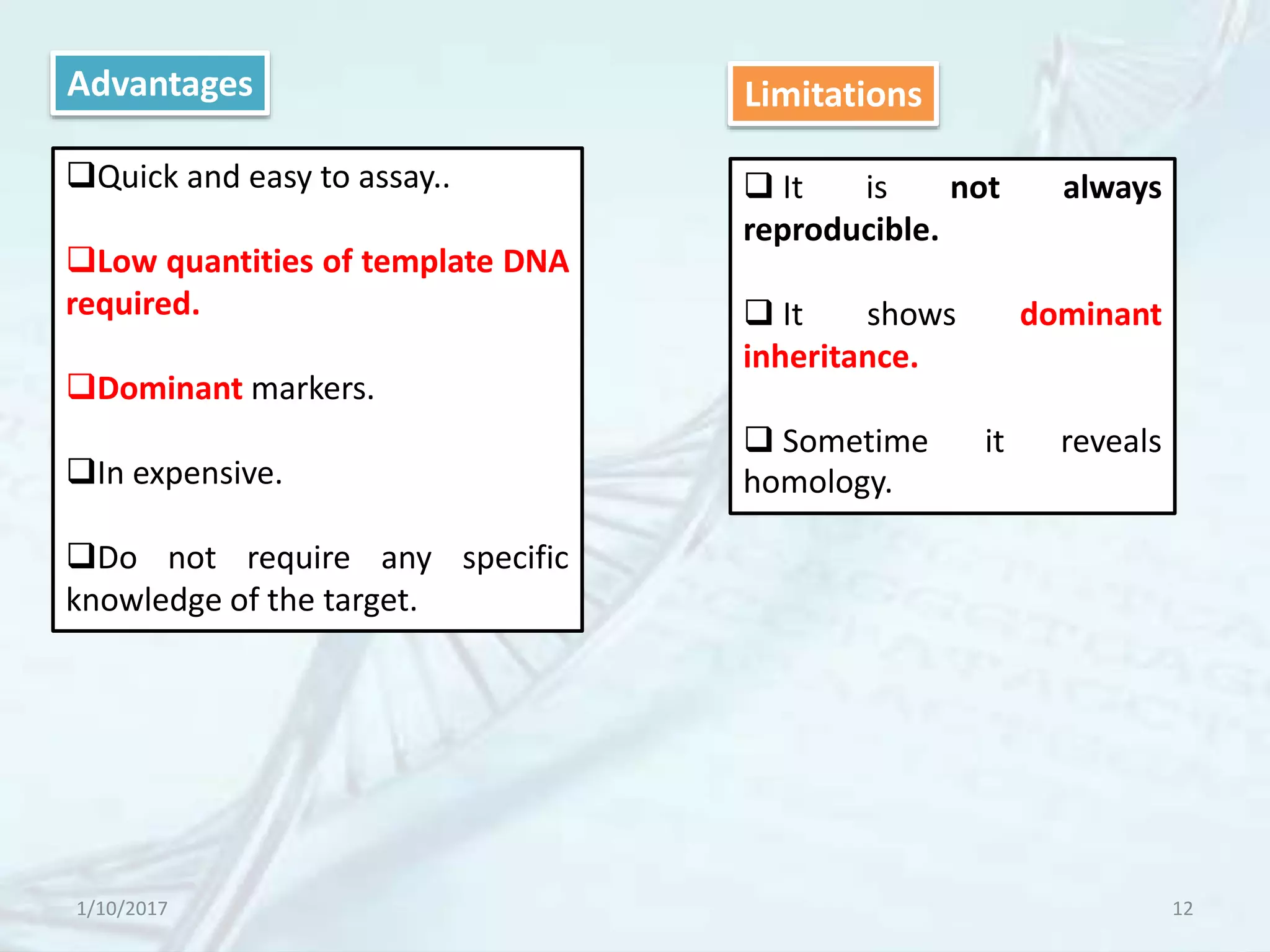
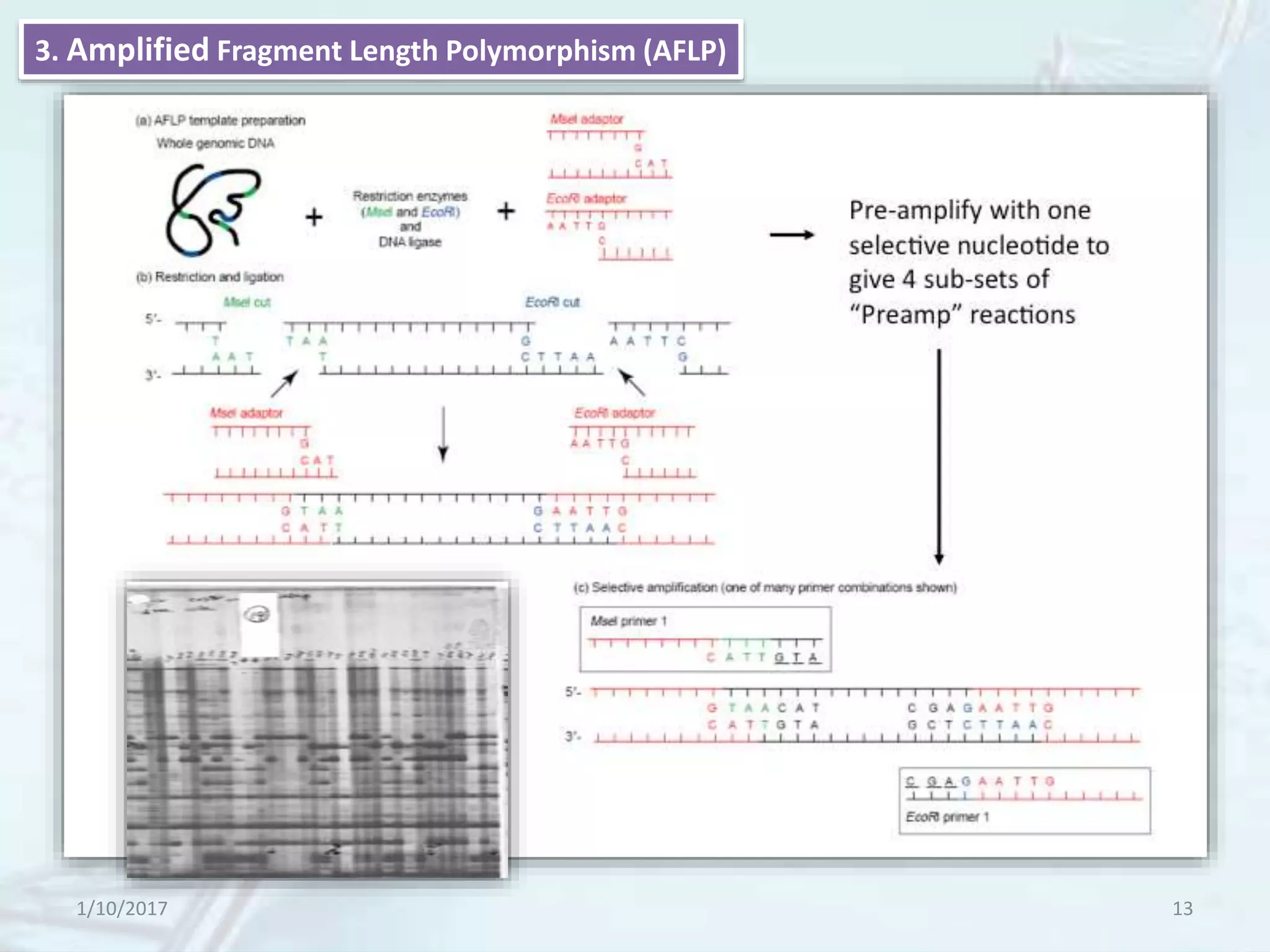
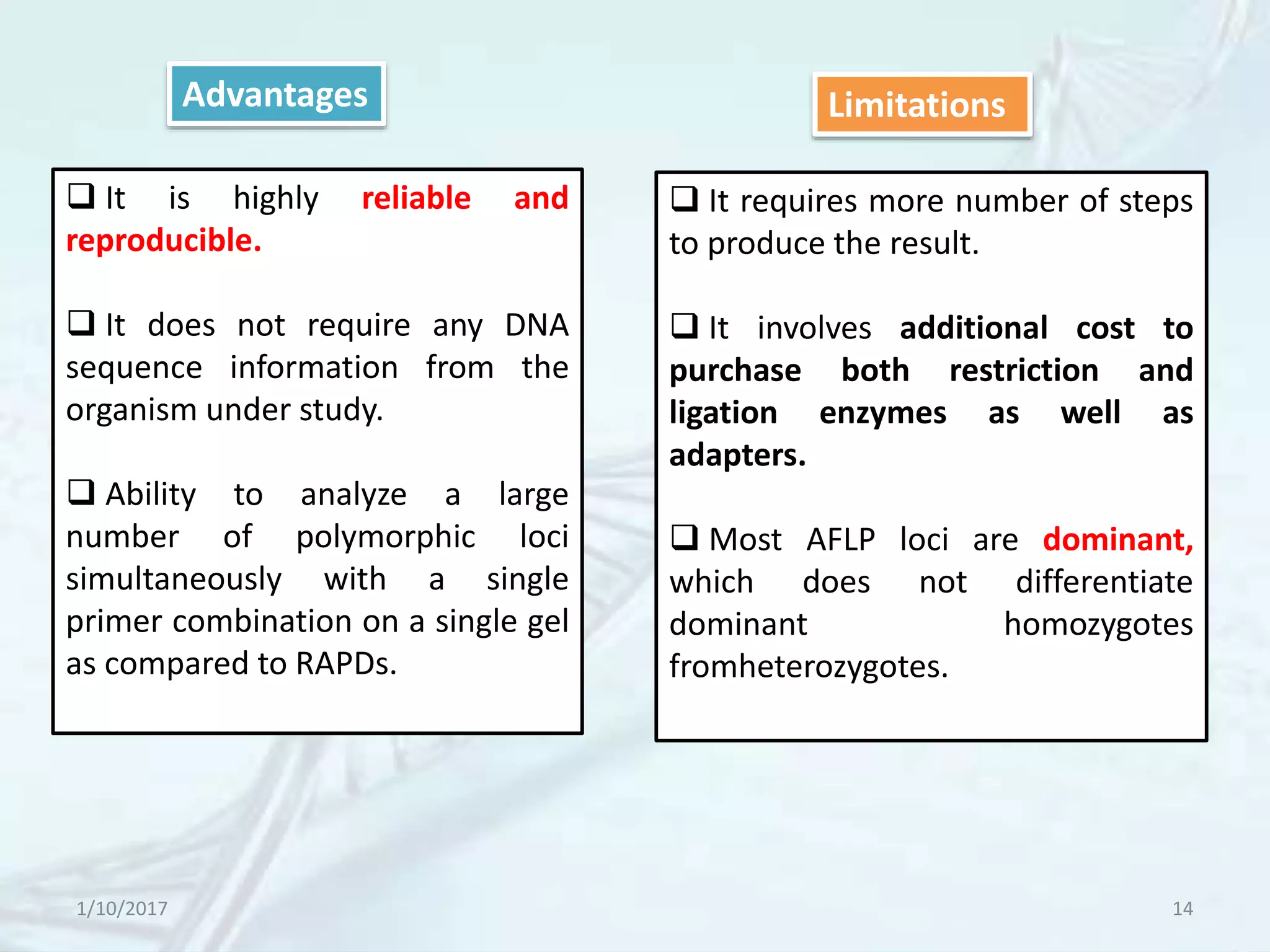
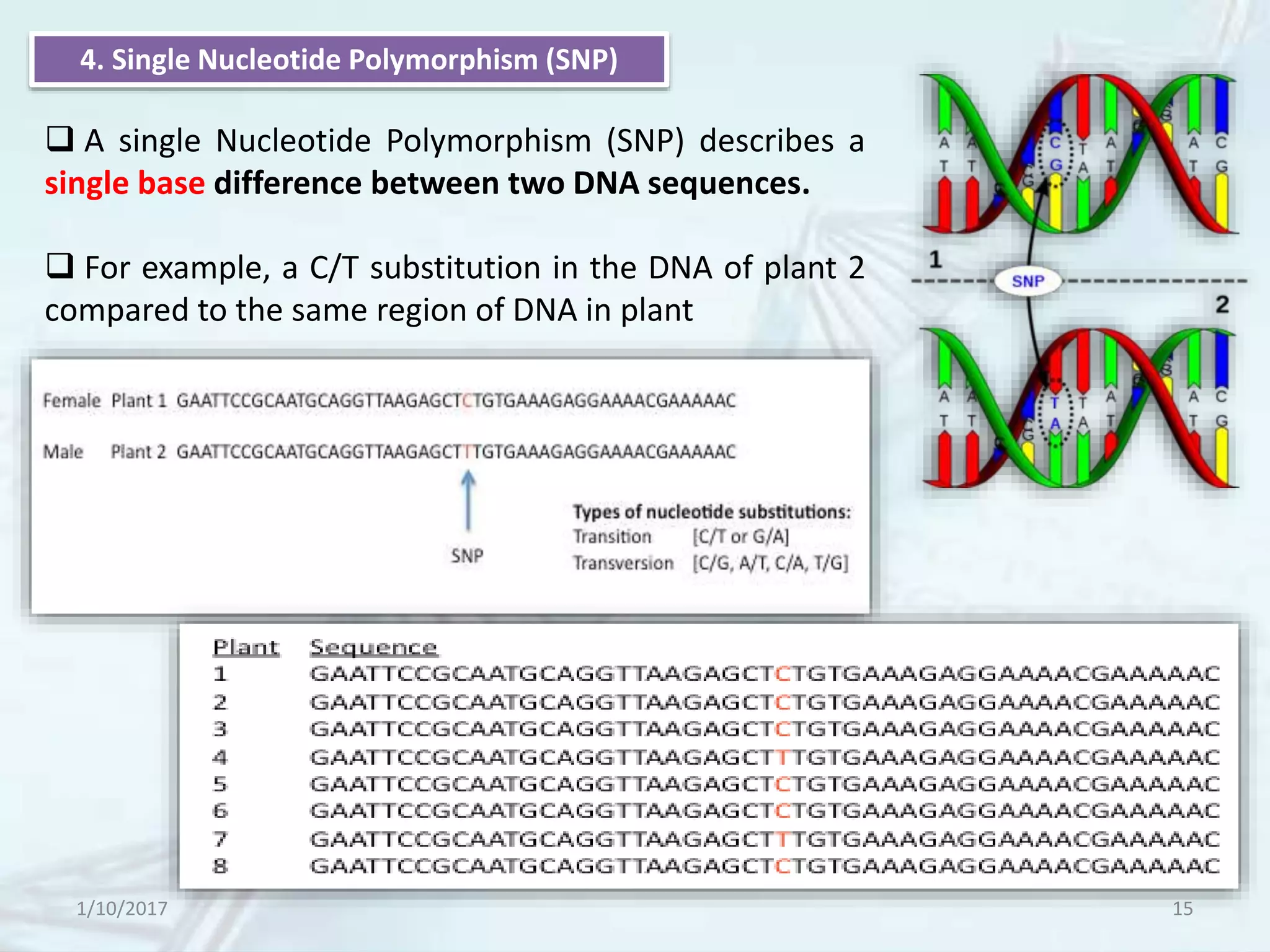
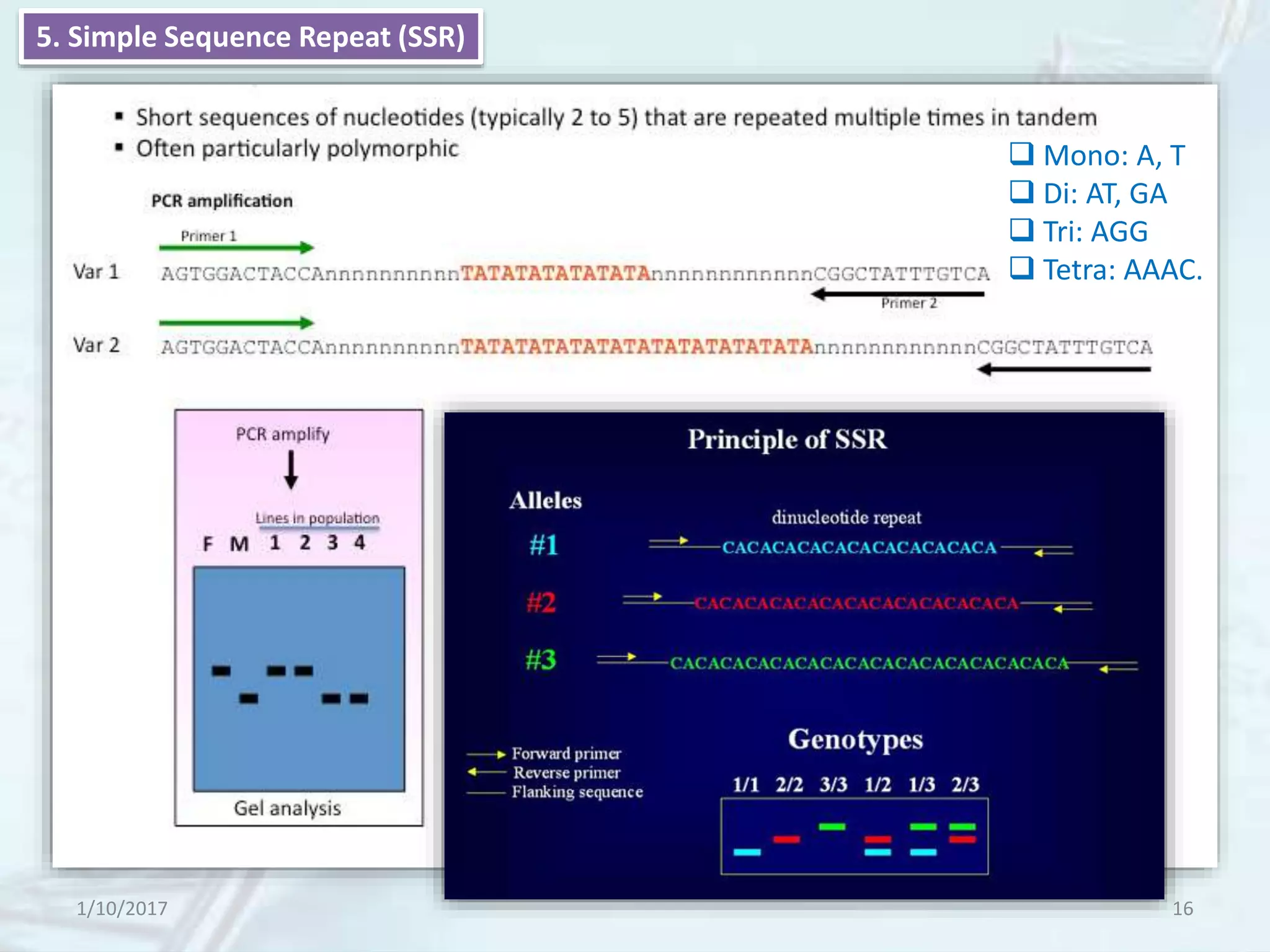
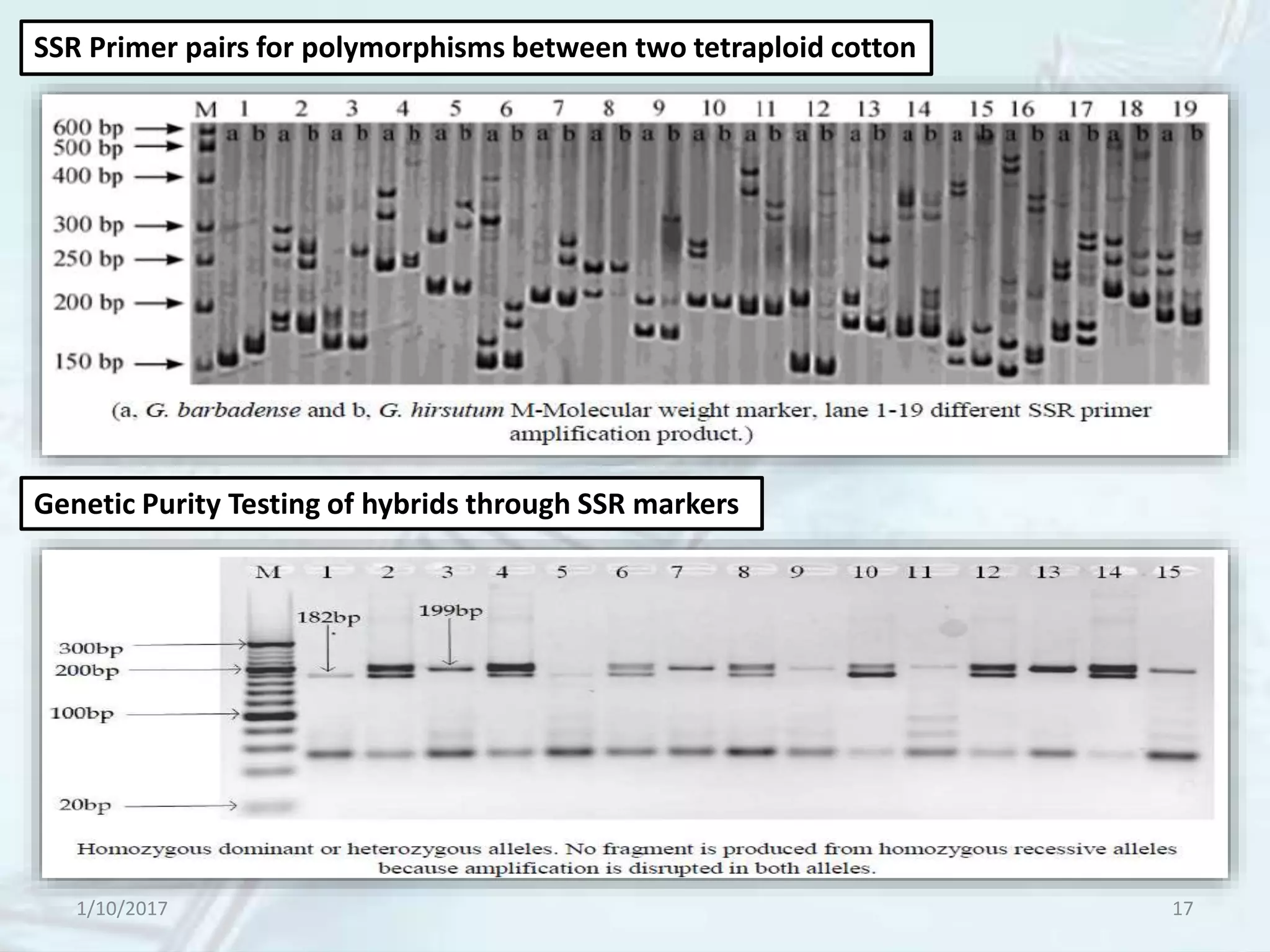
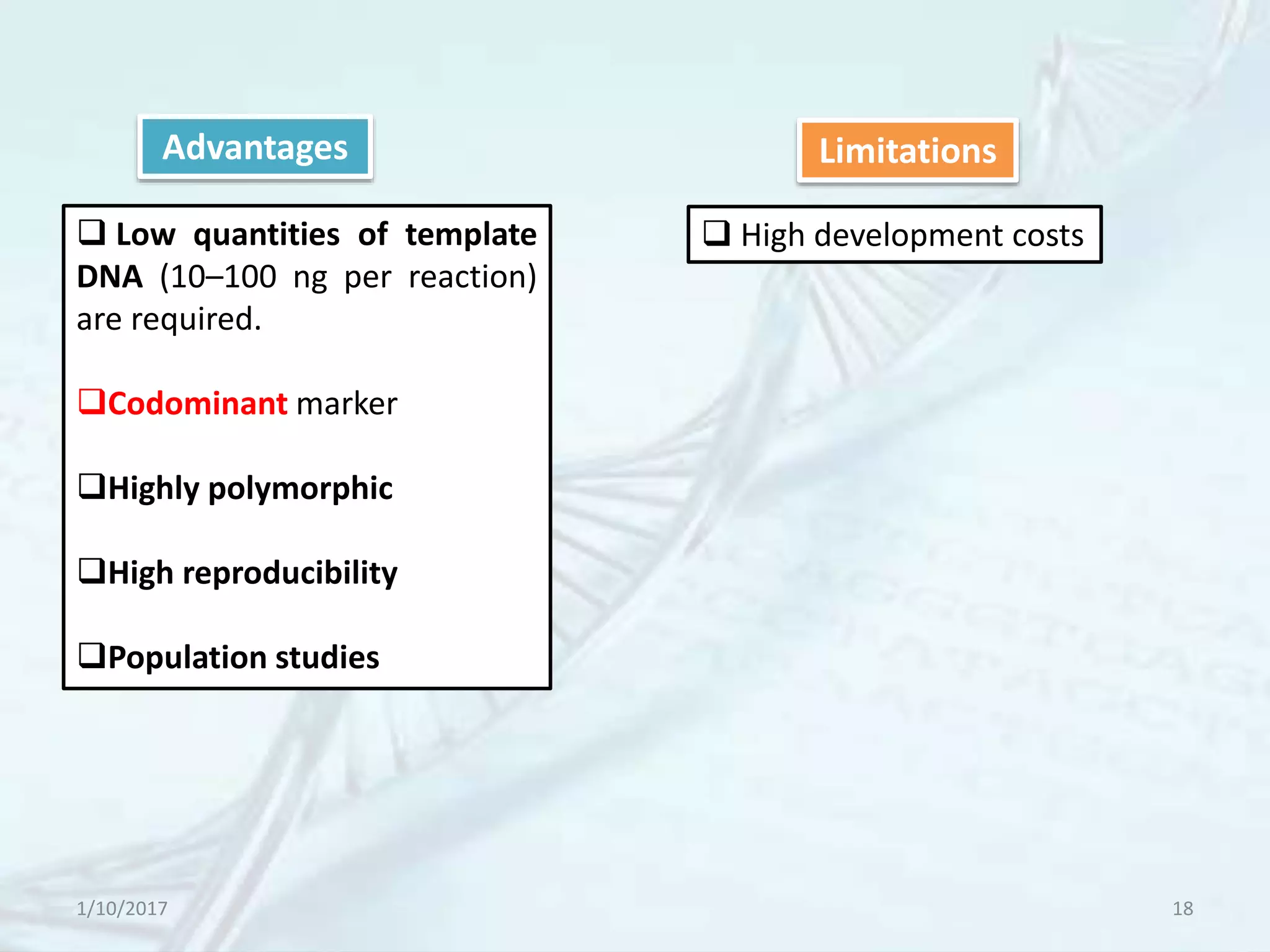
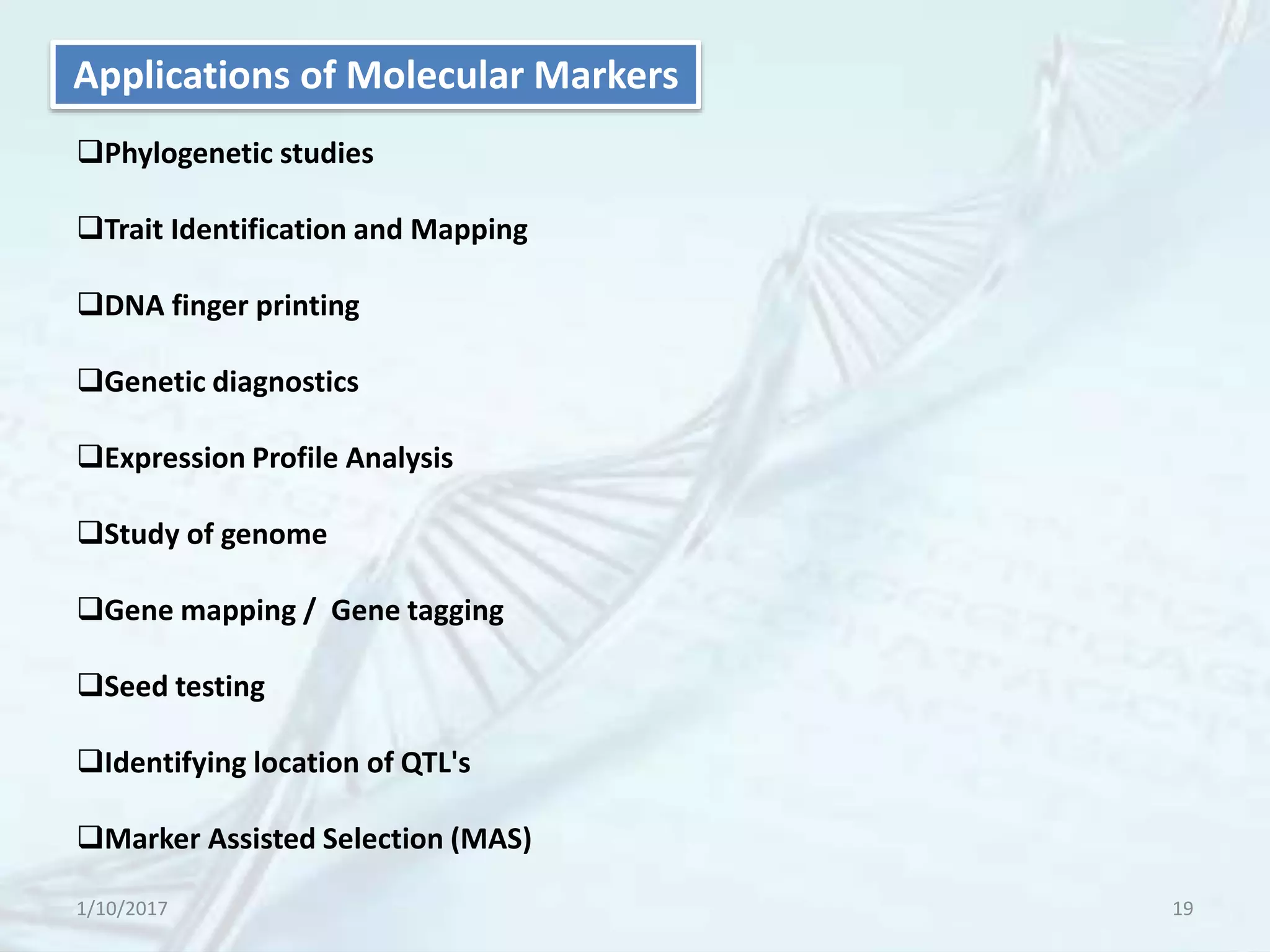
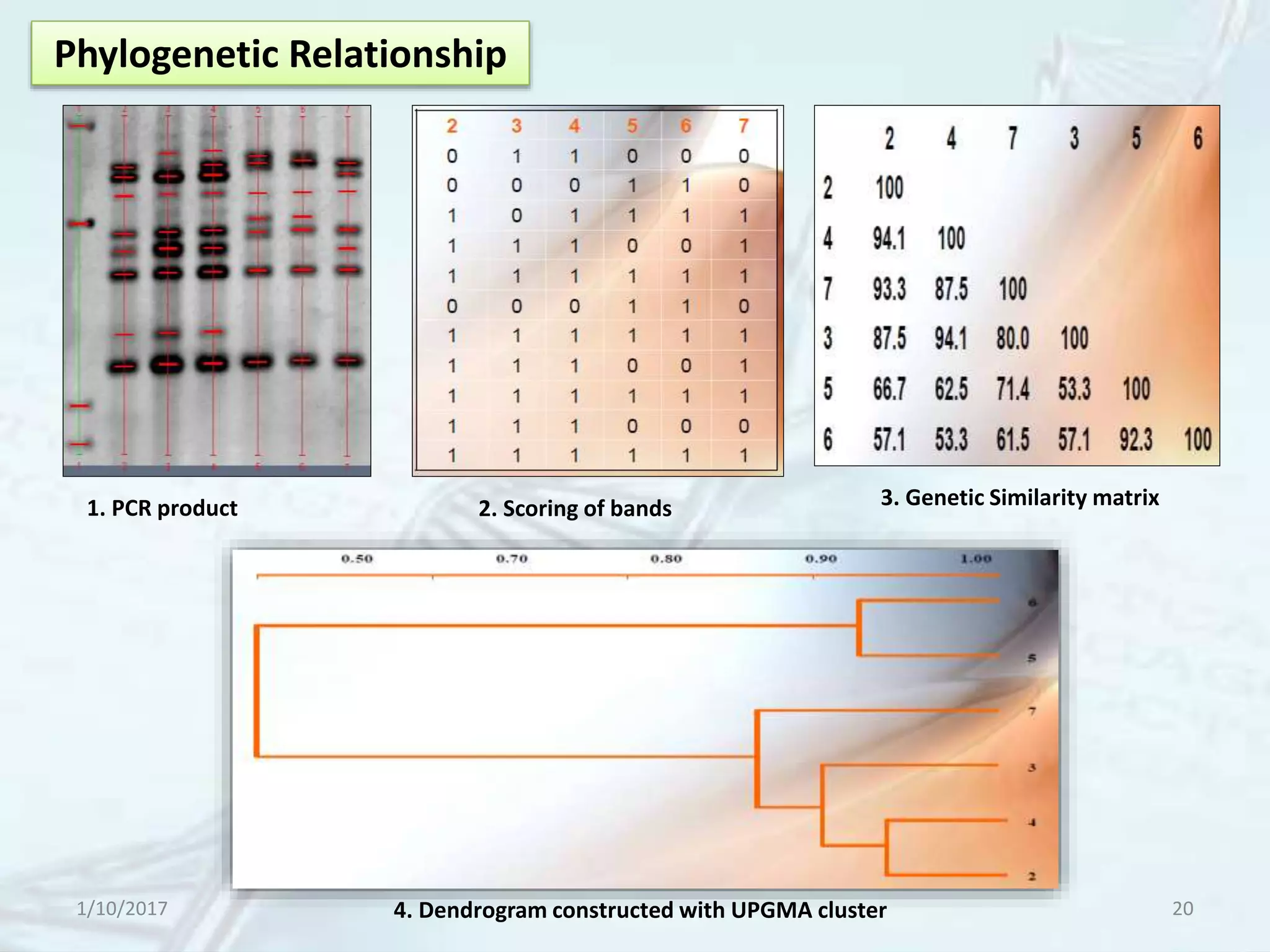
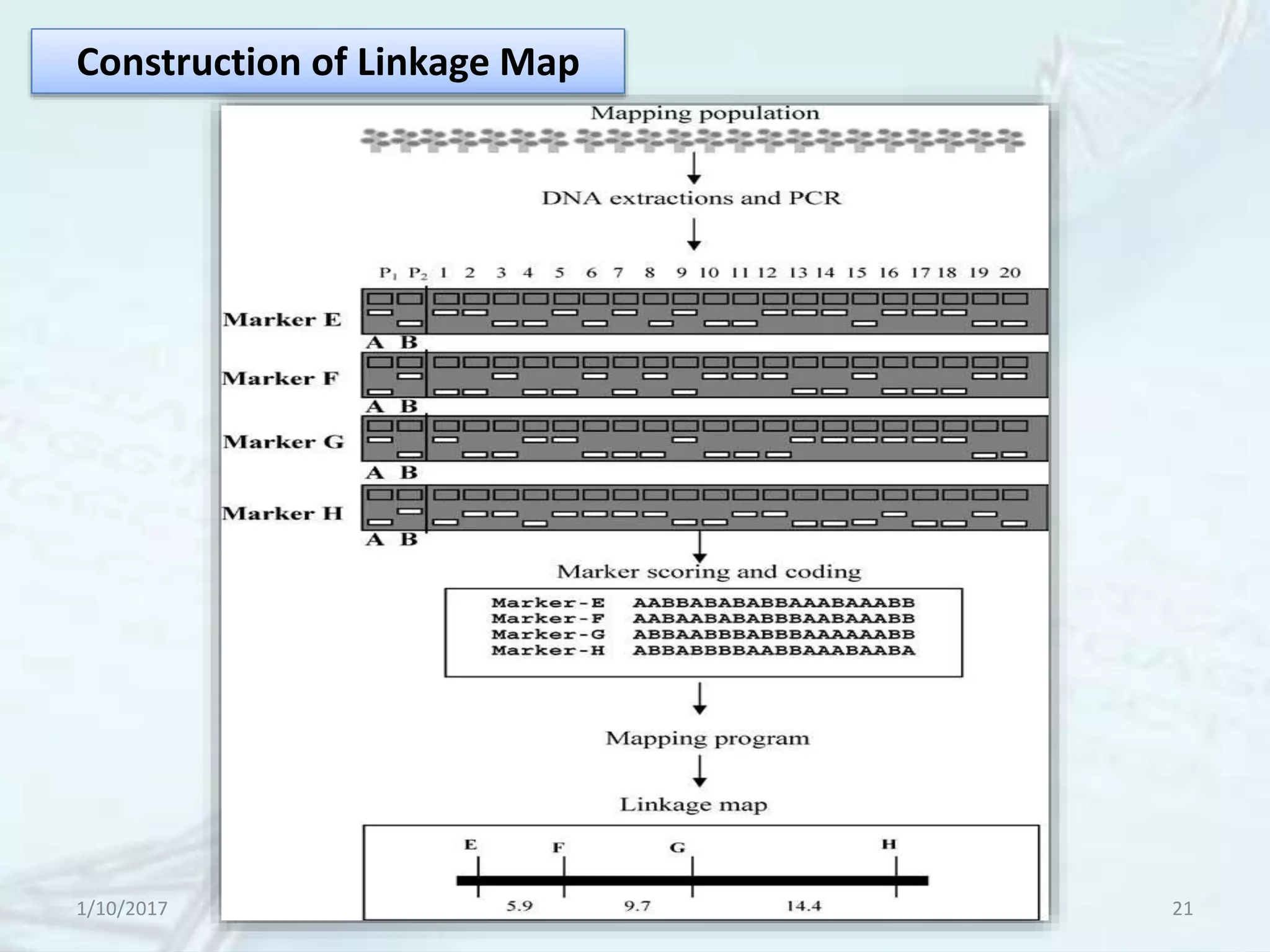
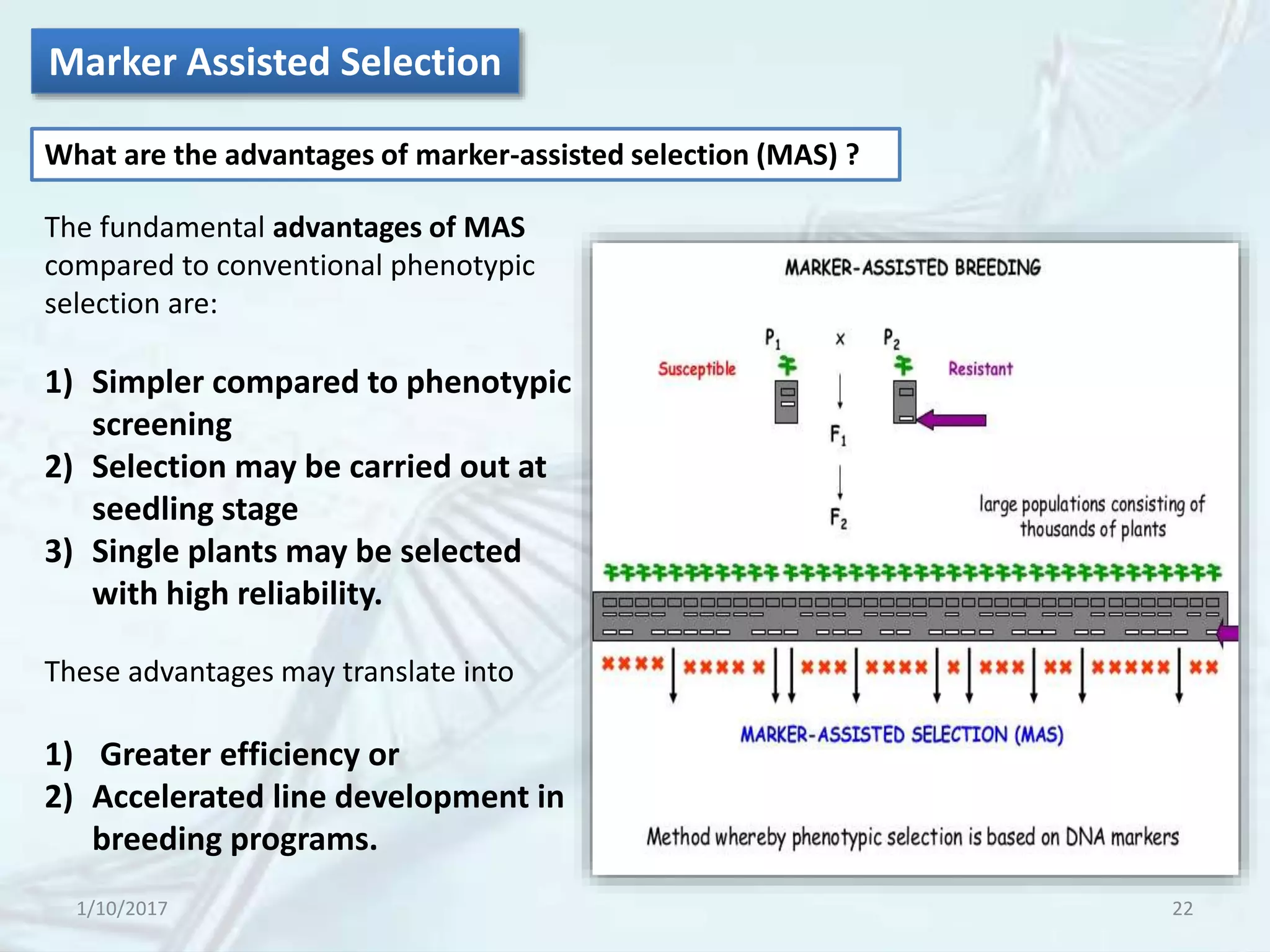
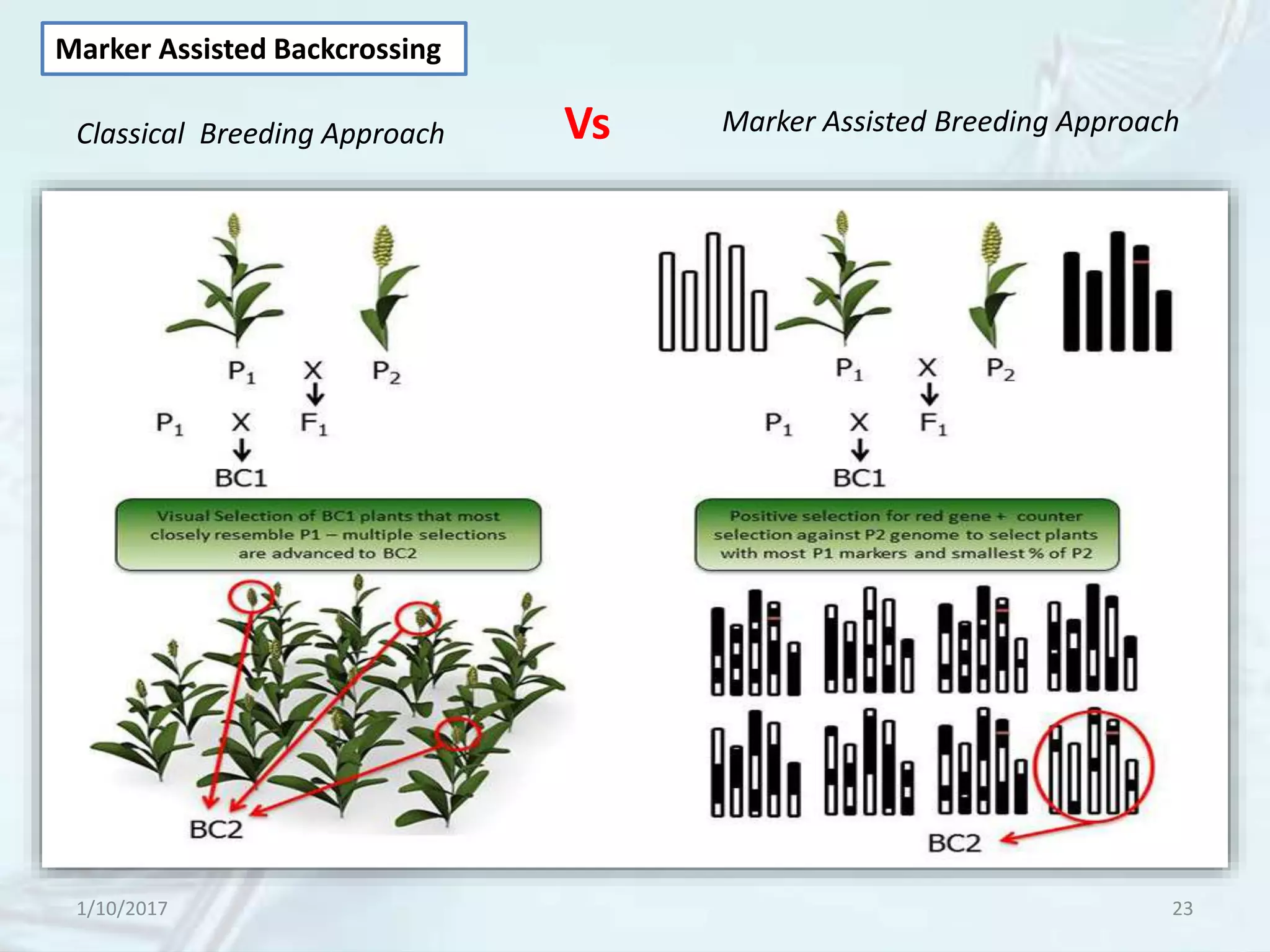
The document provides an overview of genetic markers, defining them as genes or DNA sequences located on chromosomes that serve as indicators for individuals or species. It discusses various types of genetic markers, including molecular markers, their advantages and limitations, and applications in fields such as breeding and genetic mapping. Additionally, it emphasizes the benefits of marker-assisted selection (MAS) over traditional phenotypic selection in breeding programs.