This document provides an overview of fluorescence in situ hybridization (FISH). It discusses:
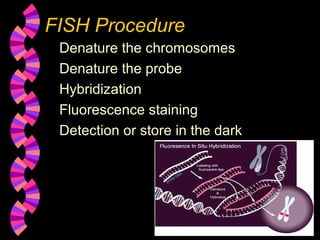
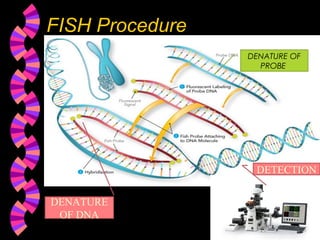
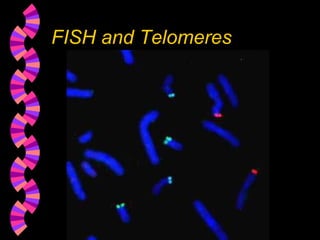
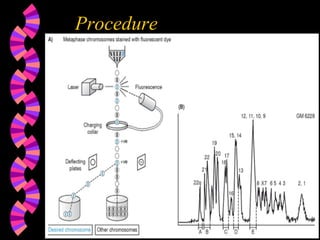
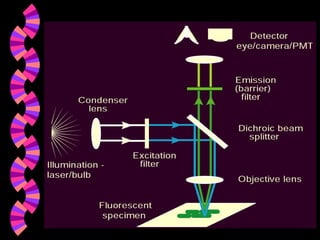
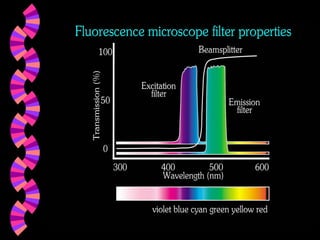
1. FISH is a cytogenetic technique that uses fluorescent probes to bind specifically to chromosomal regions with complementary DNA sequences, which can then be viewed under a fluorescent microscope.
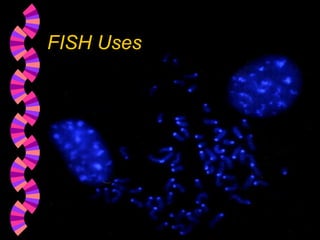
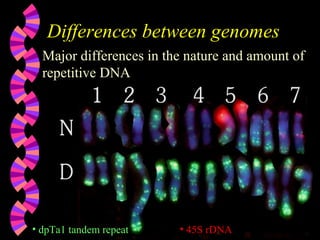
2. FISH can be used to identify chromosomal abnormalities, assist with gene mapping, and determine ploidy. It has advantages over other techniques like being less labor intensive.
3. Common applications of FISH include detecting aneuploidies for prenatal diagnosis, locating specific DNA sequences on chromosomes, and identifying features in DNA for uses like genetic counseling.