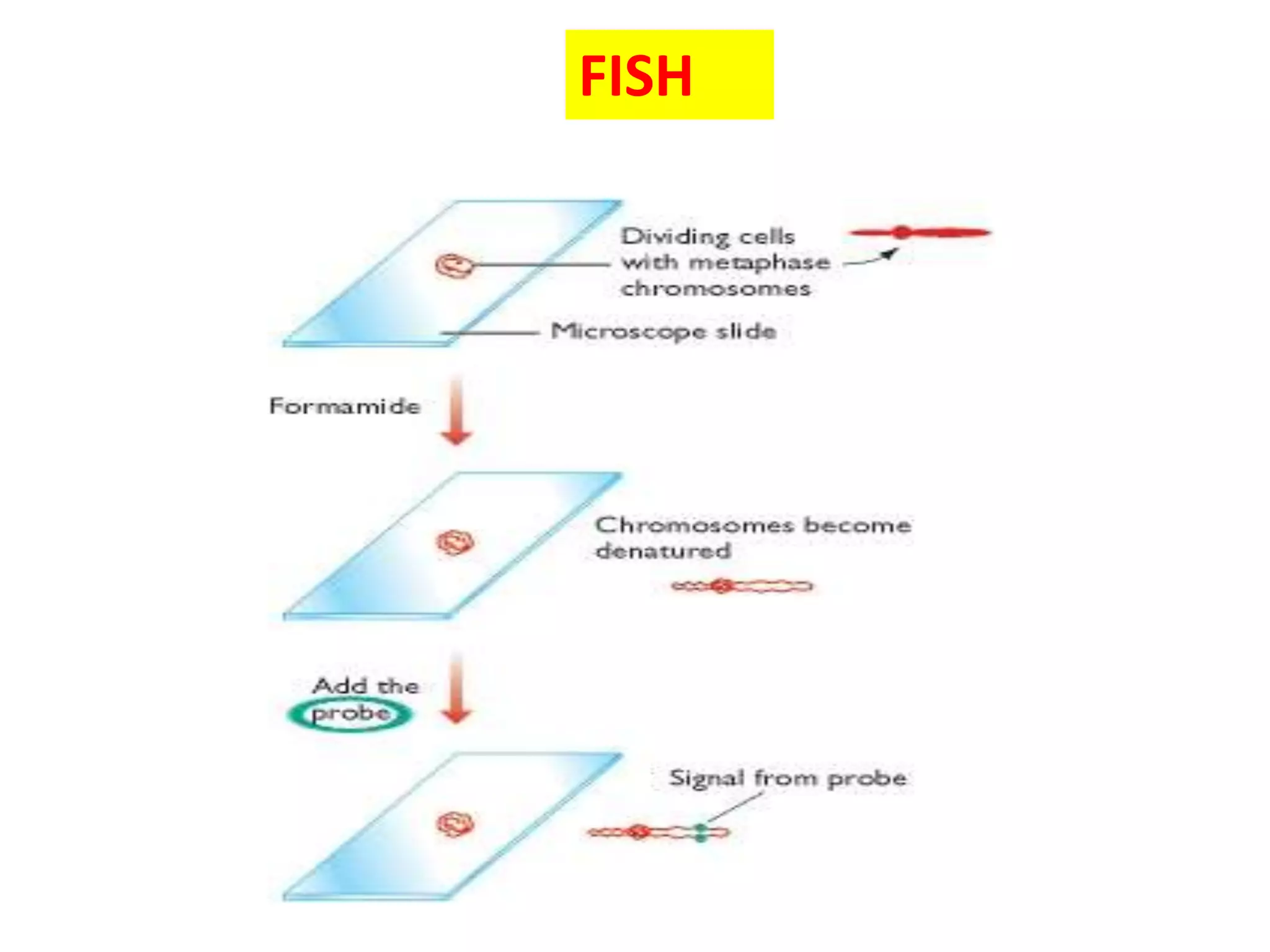
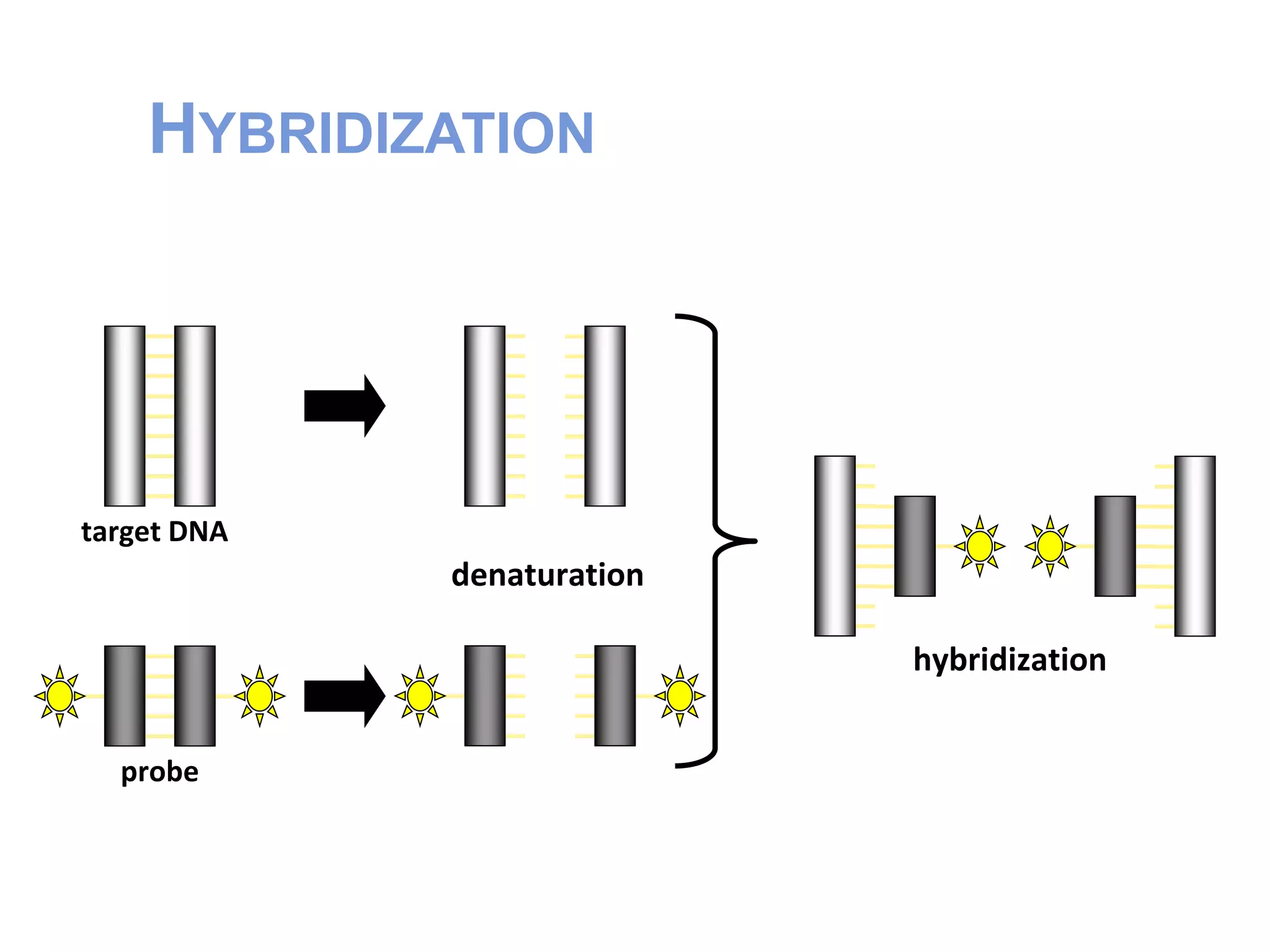
FISH (Fluorescent In-Situ Hybridization) is a technique that uses fluorescent probes that bind to complementary DNA sequences to localize specific genes or DNA sequences on chromosomes. It allows for direct visualization of the position of genetic markers on chromosomes. The FISH procedure involves denaturing chromosomes and probes, hybridizing the probes to target sequences, and examining slides under a fluorescence microscope. FISH has advantages like being rapid and allowing analysis of many cells, and has been improved over time, though accurate quantification remains challenging.