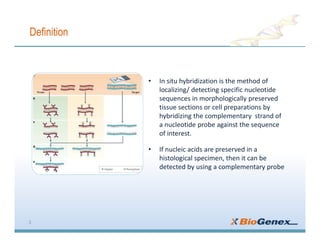
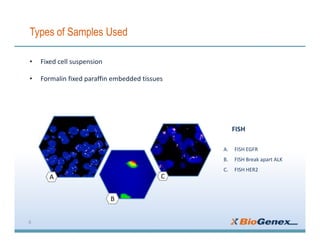
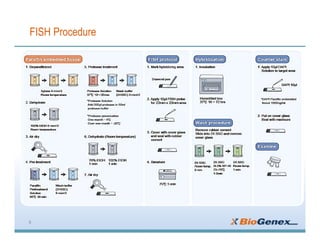
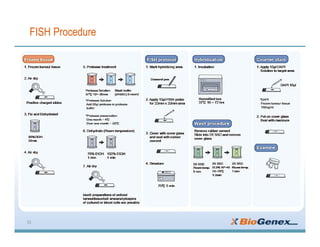
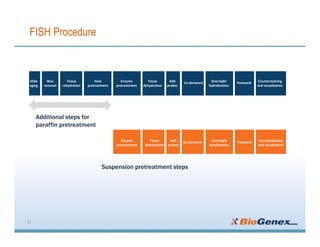
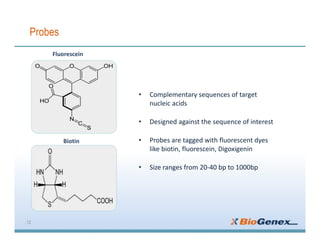
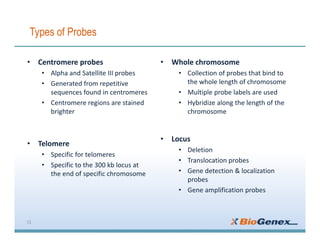
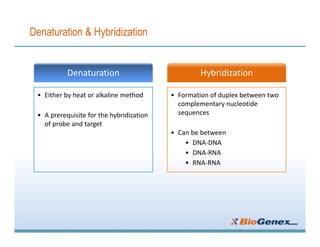
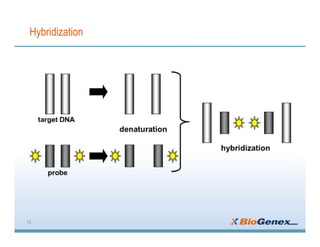
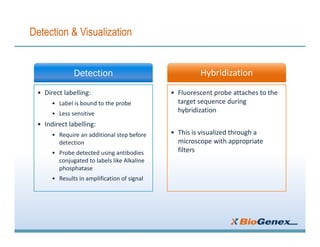
Fluorescent in-situ hybridization (FISH) is a technique that uses fluorescent probes that bind to only parts of the genome to detect and localize specific DNA sequences on chromosomes. It involves denaturing and hybridizing a fluorescent probe to a complementary DNA or RNA target sequence on preserved tissue. The bound probe can then be visualized with a fluorescent microscope to identify specific gene mutations, abnormalities, or other chromosome changes to aid in cancer diagnosis, prenatal testing, and other clinical applications.