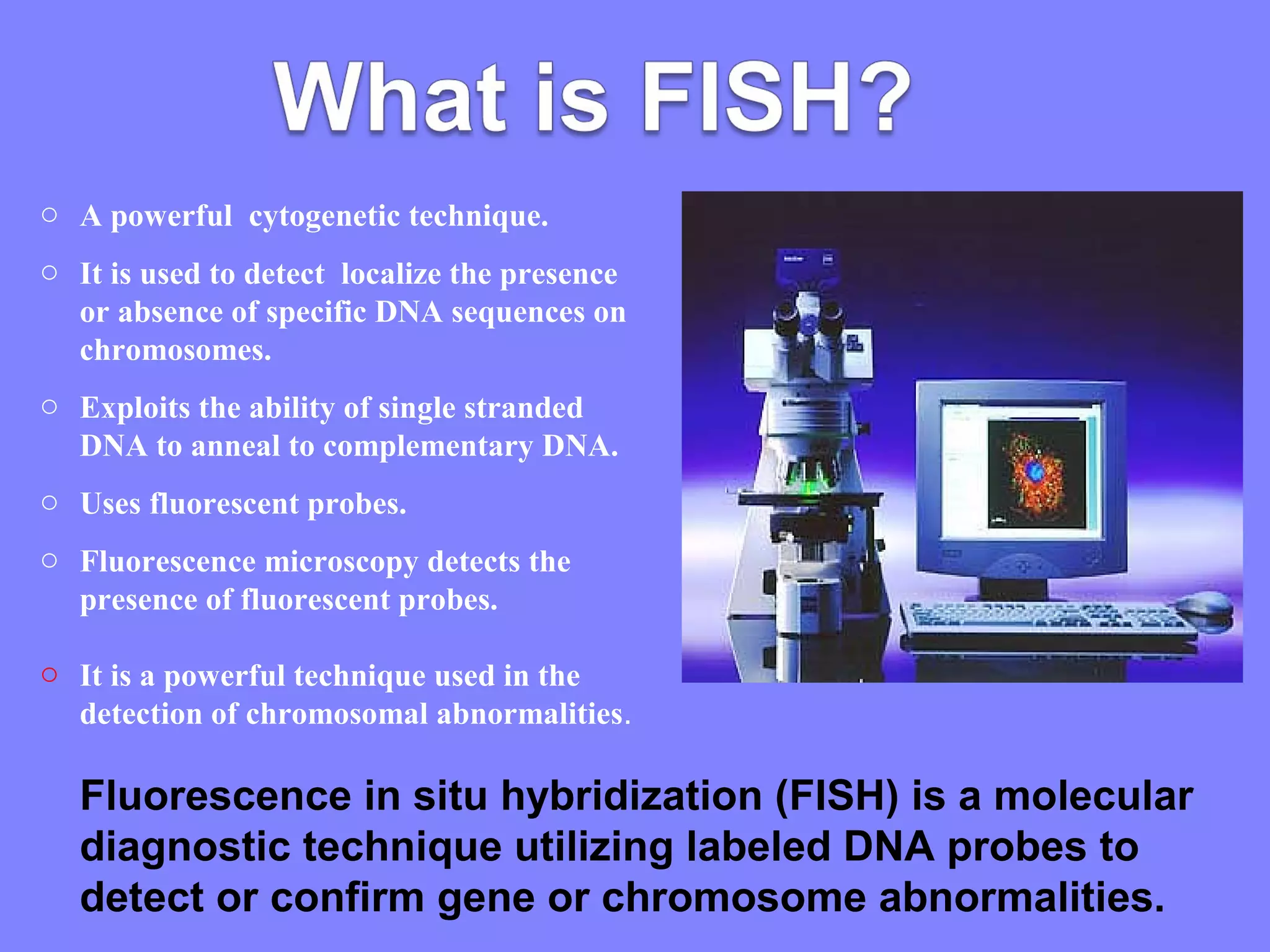
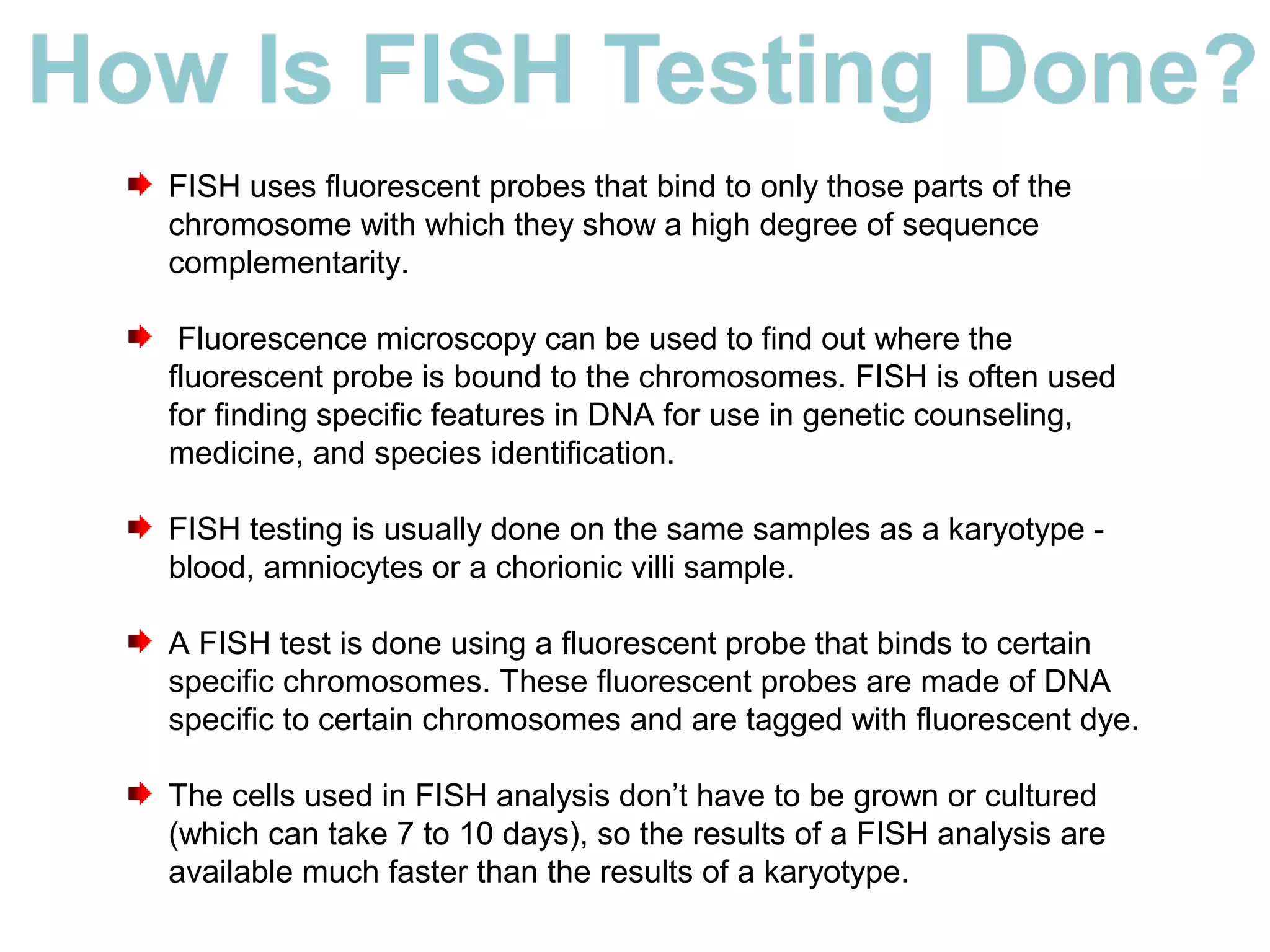
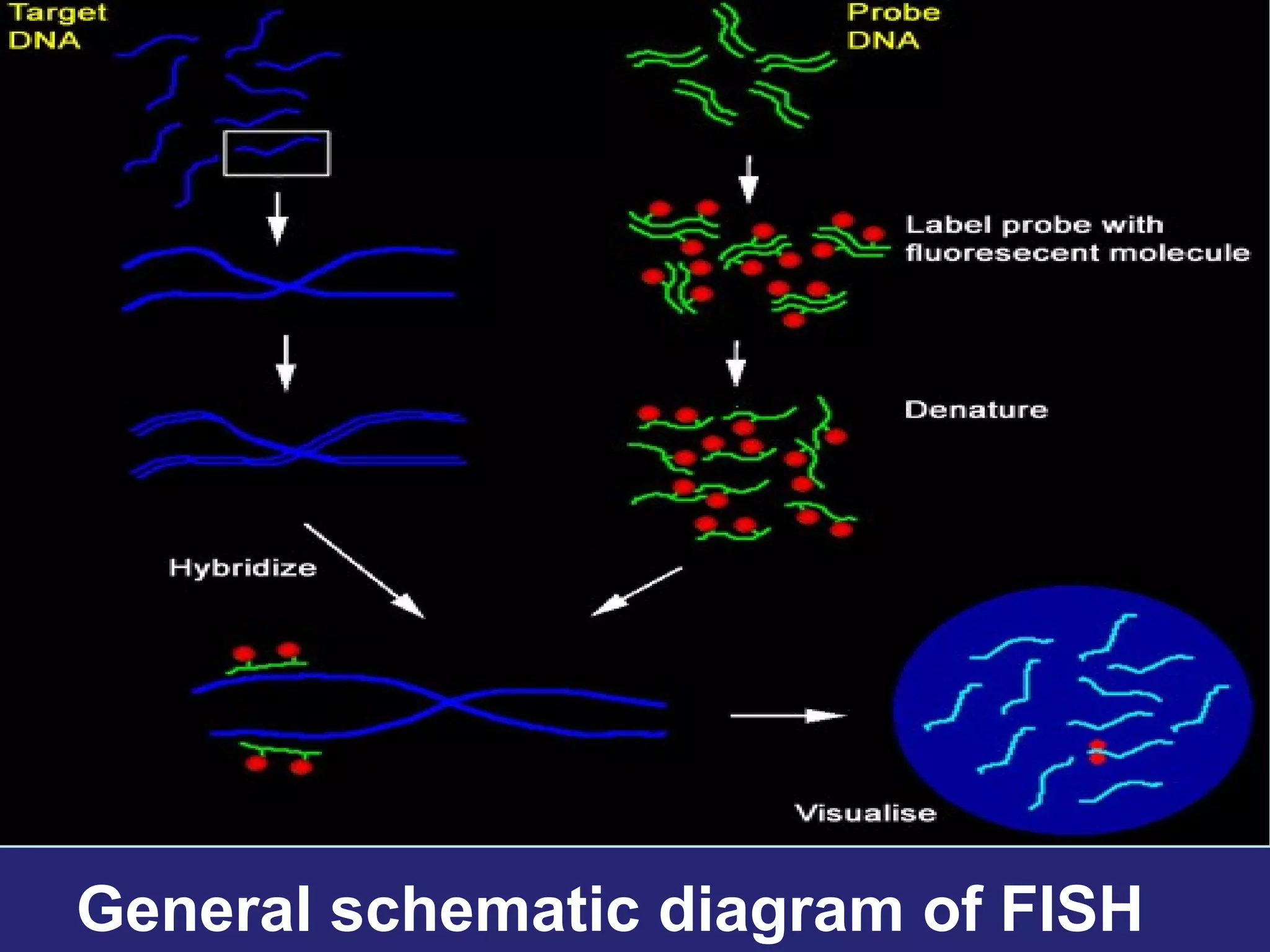
Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only parts of the chromosome with high sequence complementarity. It allows researchers to detect and localize the presence or absence of specific DNA sequences on chromosomes or specific genes. FISH utilizes fluorescent probes and fluorescence microscopy to identify the probes' location on chromosomes to detect chromosomal abnormalities. It provides results faster than conventional karyotyping and is used to diagnose various genetic conditions and cancers.
















![o FISH is performed on a variety of specimen types such as blood, bone
marrow and paraffin-embedded tissue sections.
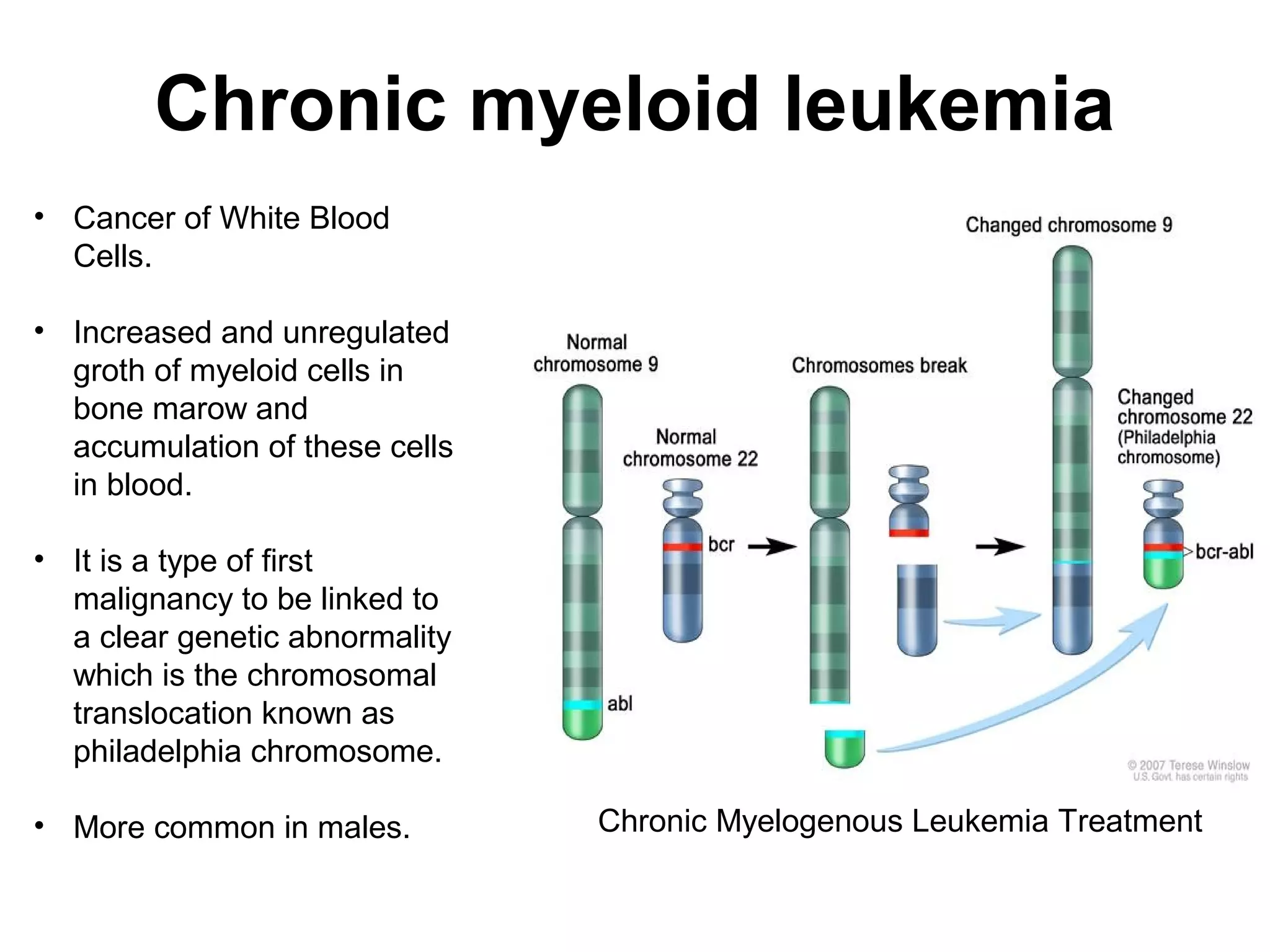
o Because of the higher sensitivity, FISH testing can detect mosaicism for
clonal chromosome changes,and is therefore useful in the detection of
minimal residual disease following treatment (e.g., interferon for chronic
myelogenous leukemia [CML]).
o Because the majority of specific deletions are sub-microscopic and can
only be detected by molecular tests, FISH plays a critical role in the
confirmation of microdeletion syndromes.
o FISH is also useful in solid tumors. For example, in determining the
prognostic role of HER-2/neu gene amplification or overexpression in
breast cancer.](https://image.slidesharecdn.com/uu-140325124838-phpapp01/75/fish-26-2048.jpg)