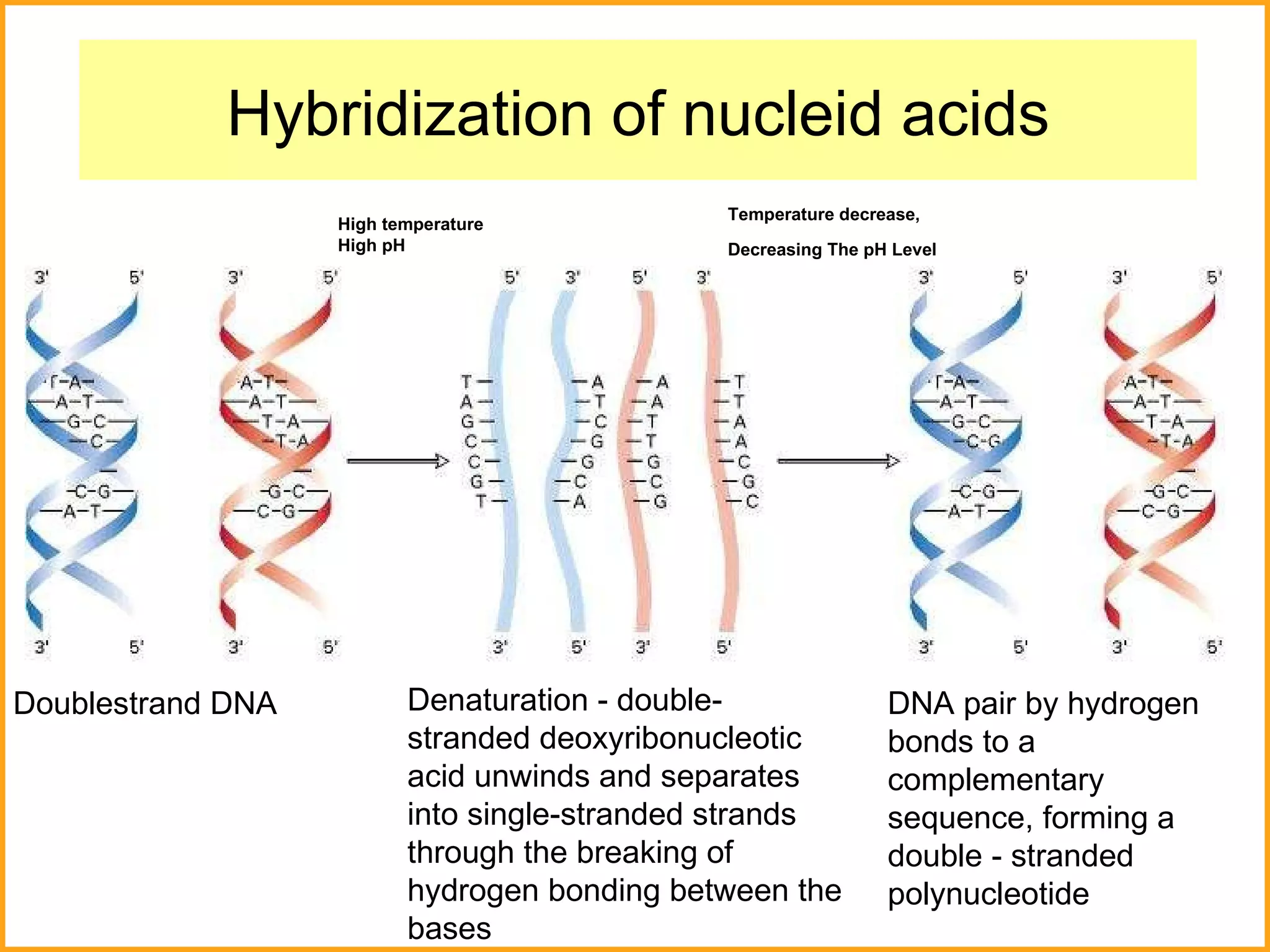
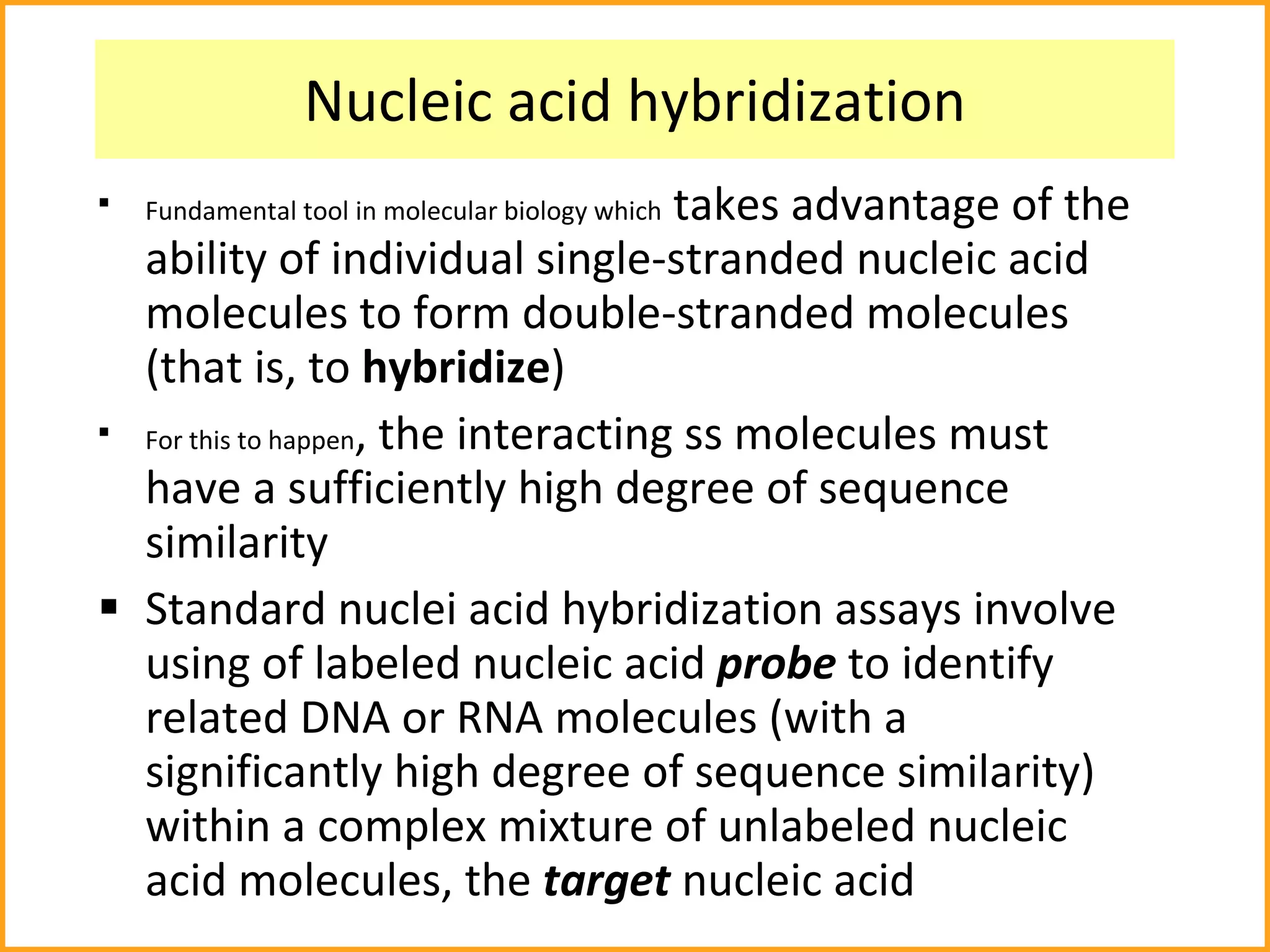
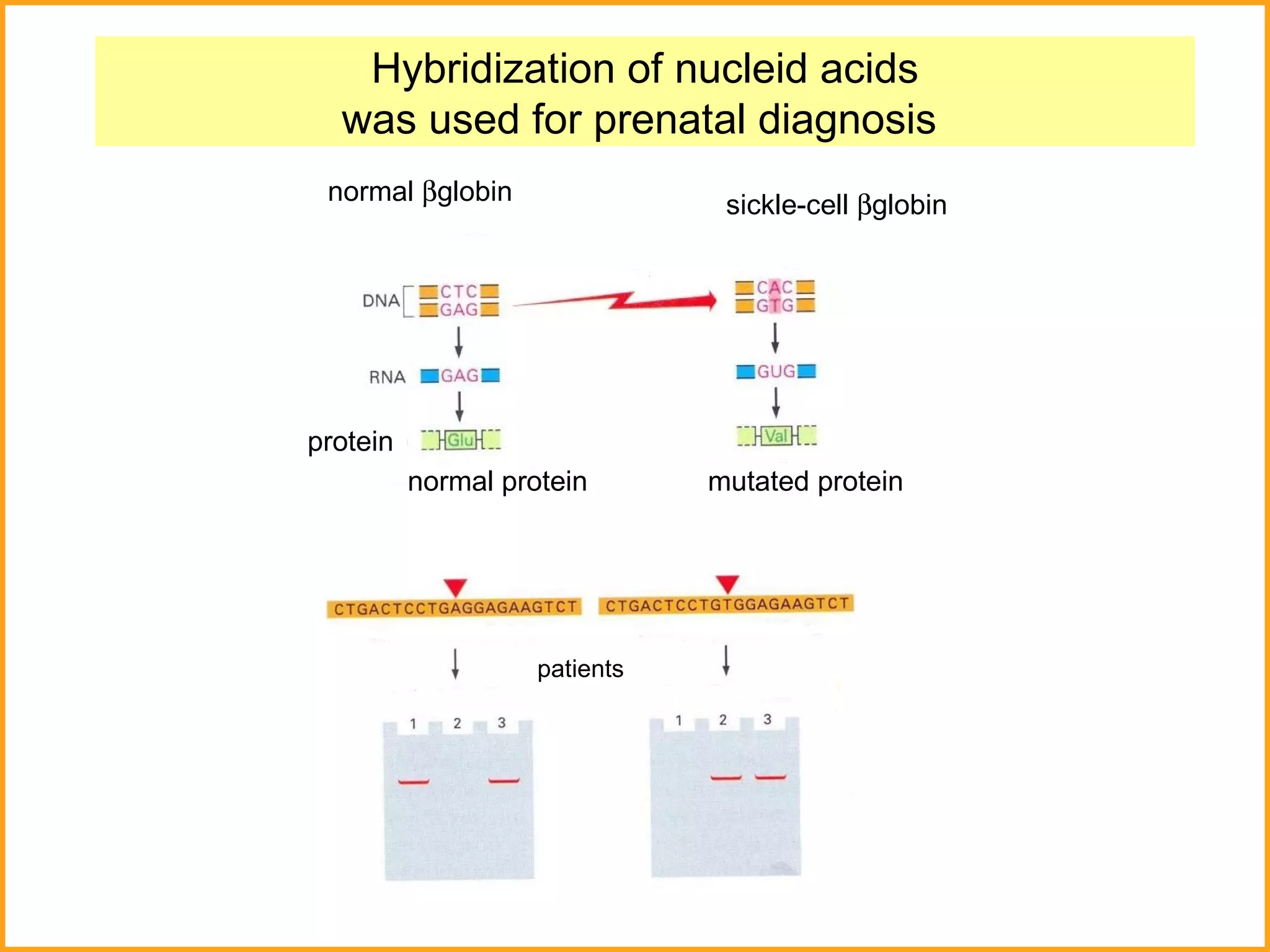
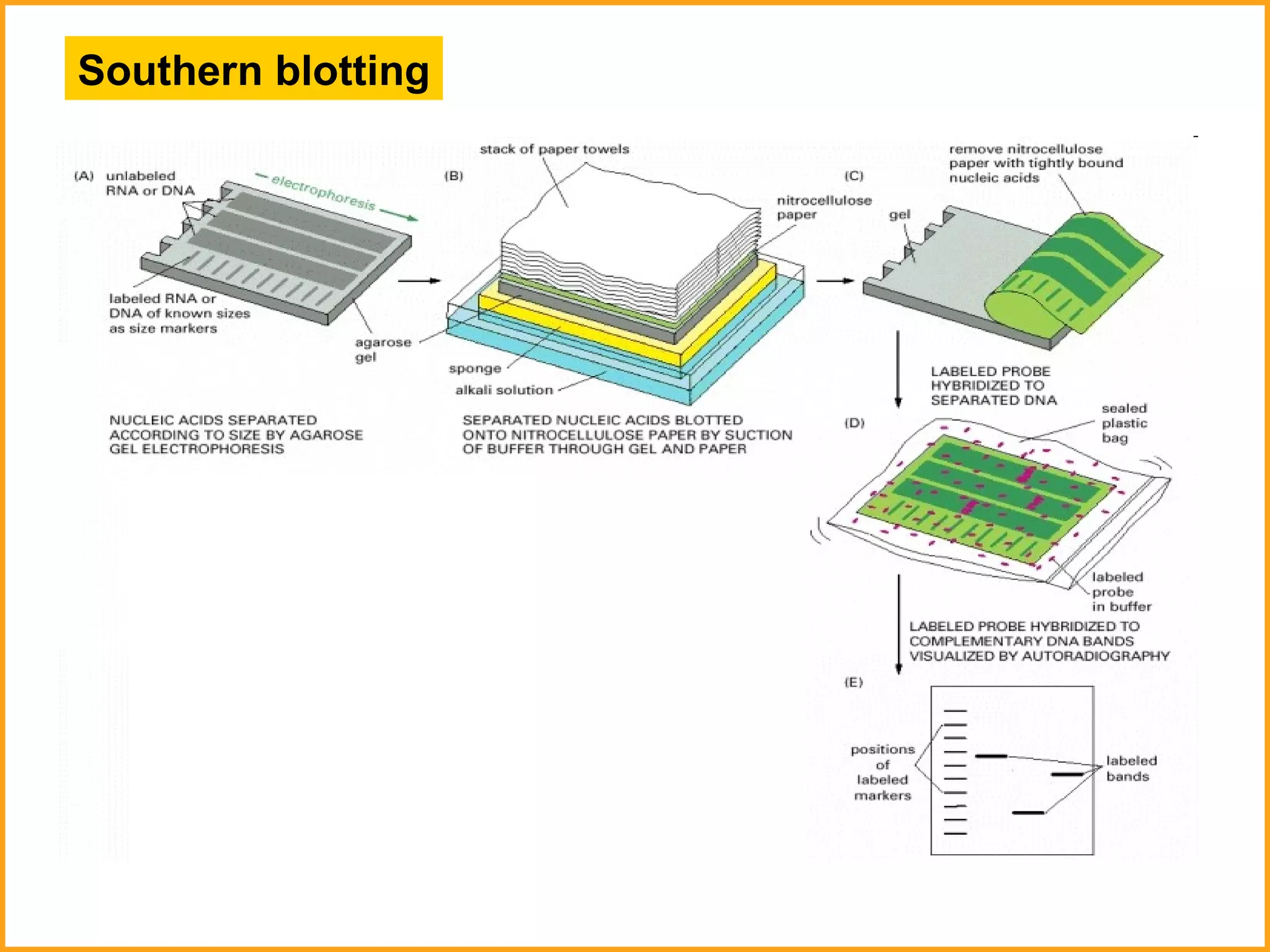
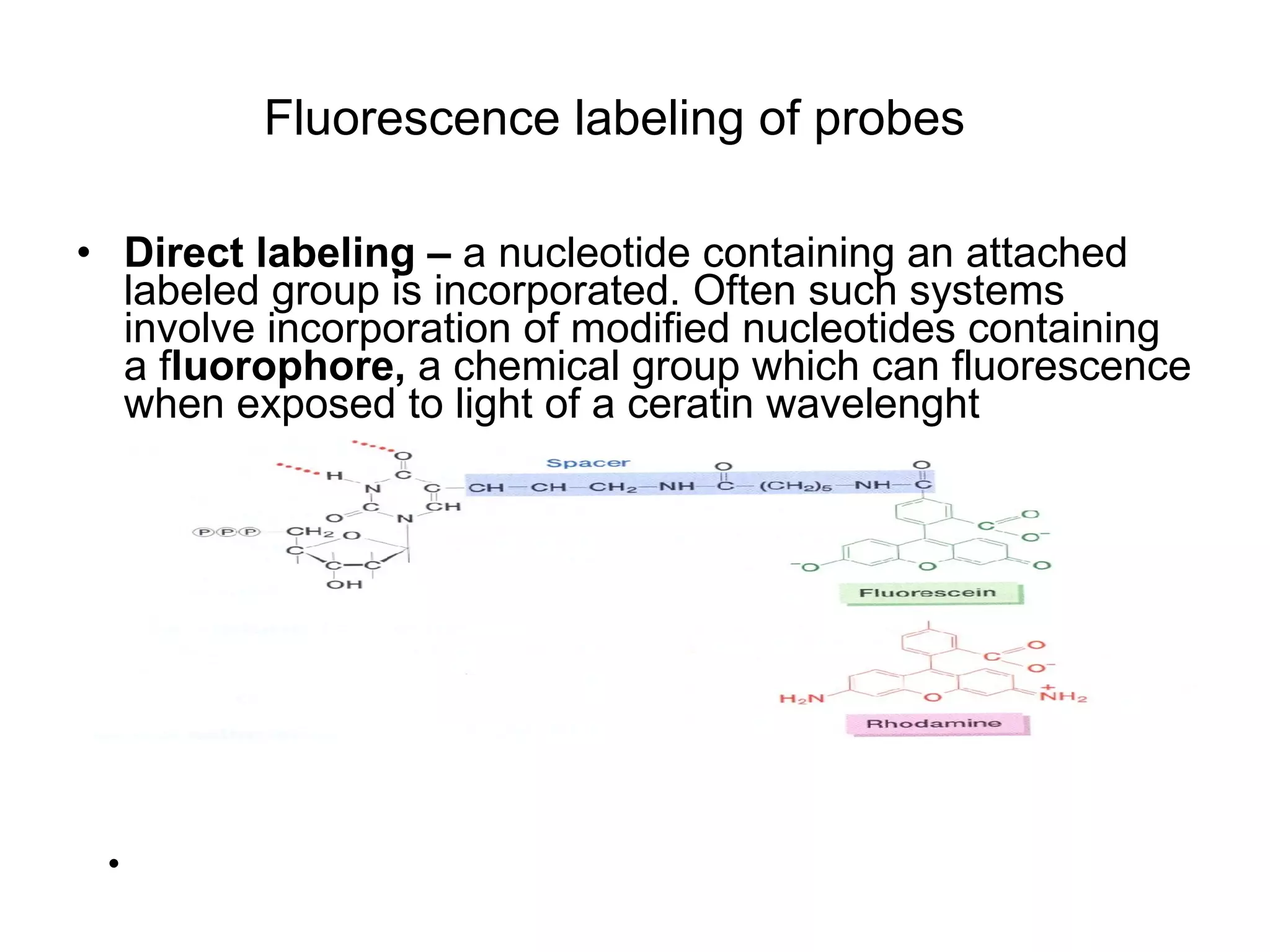
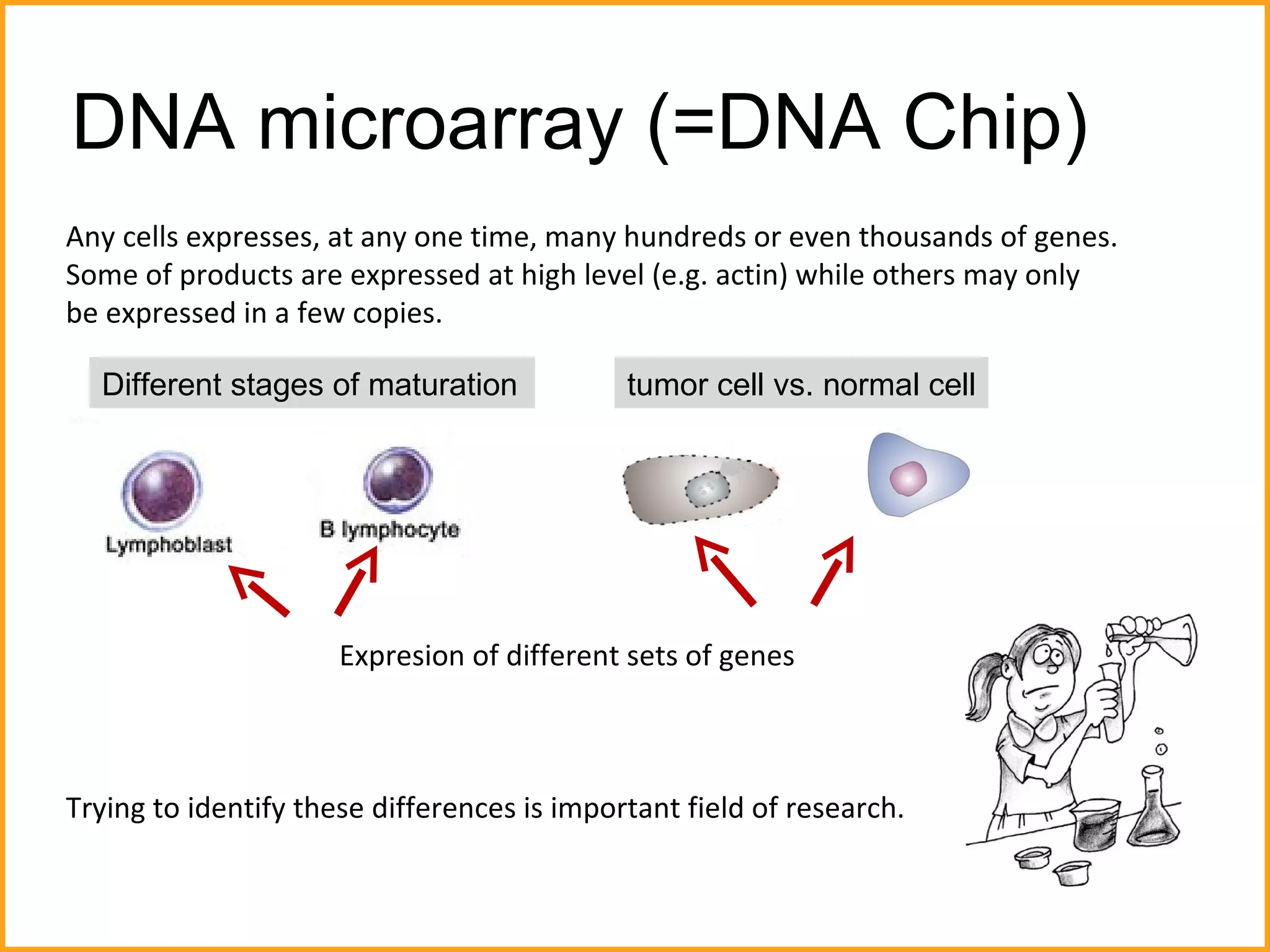
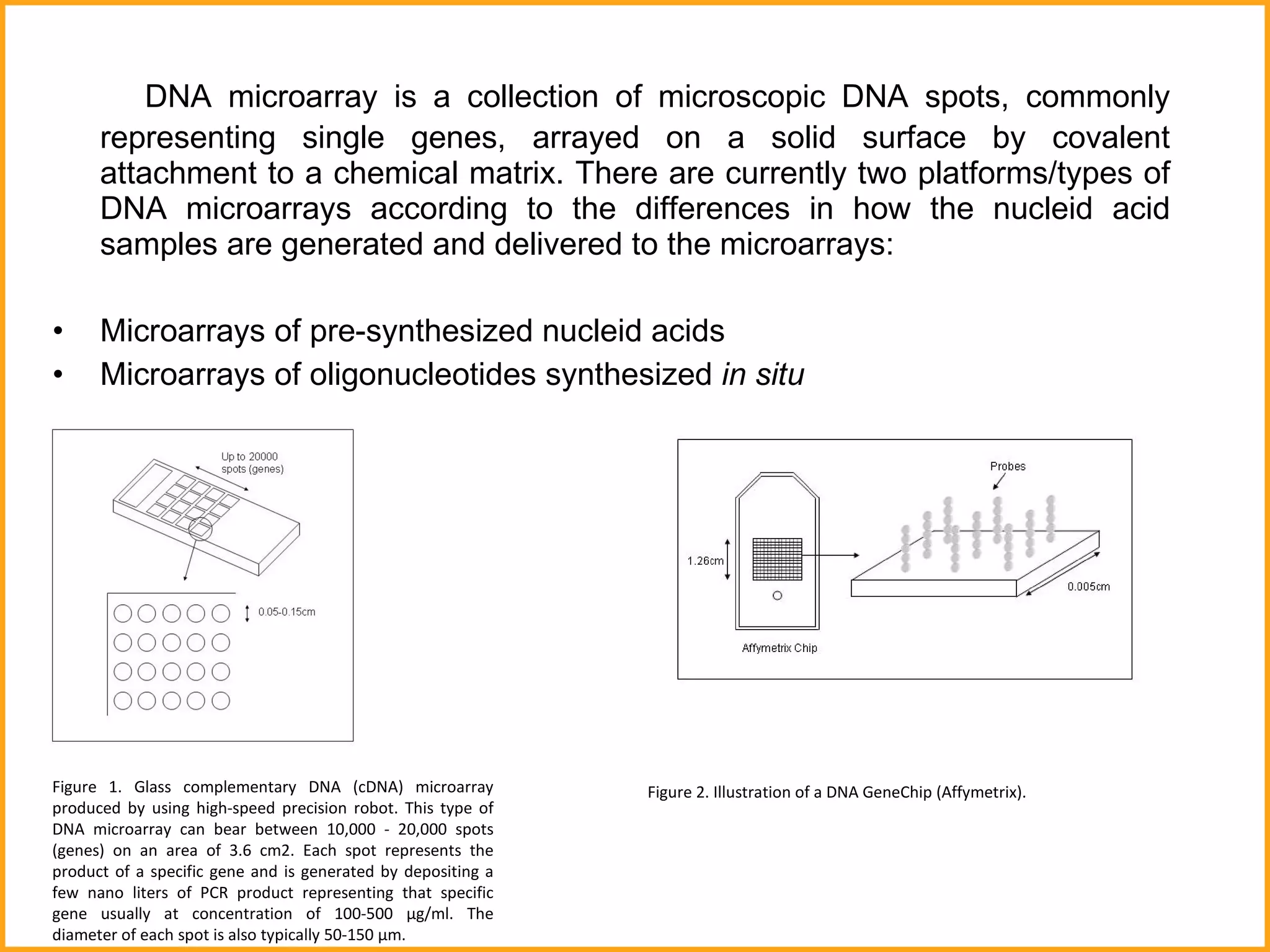
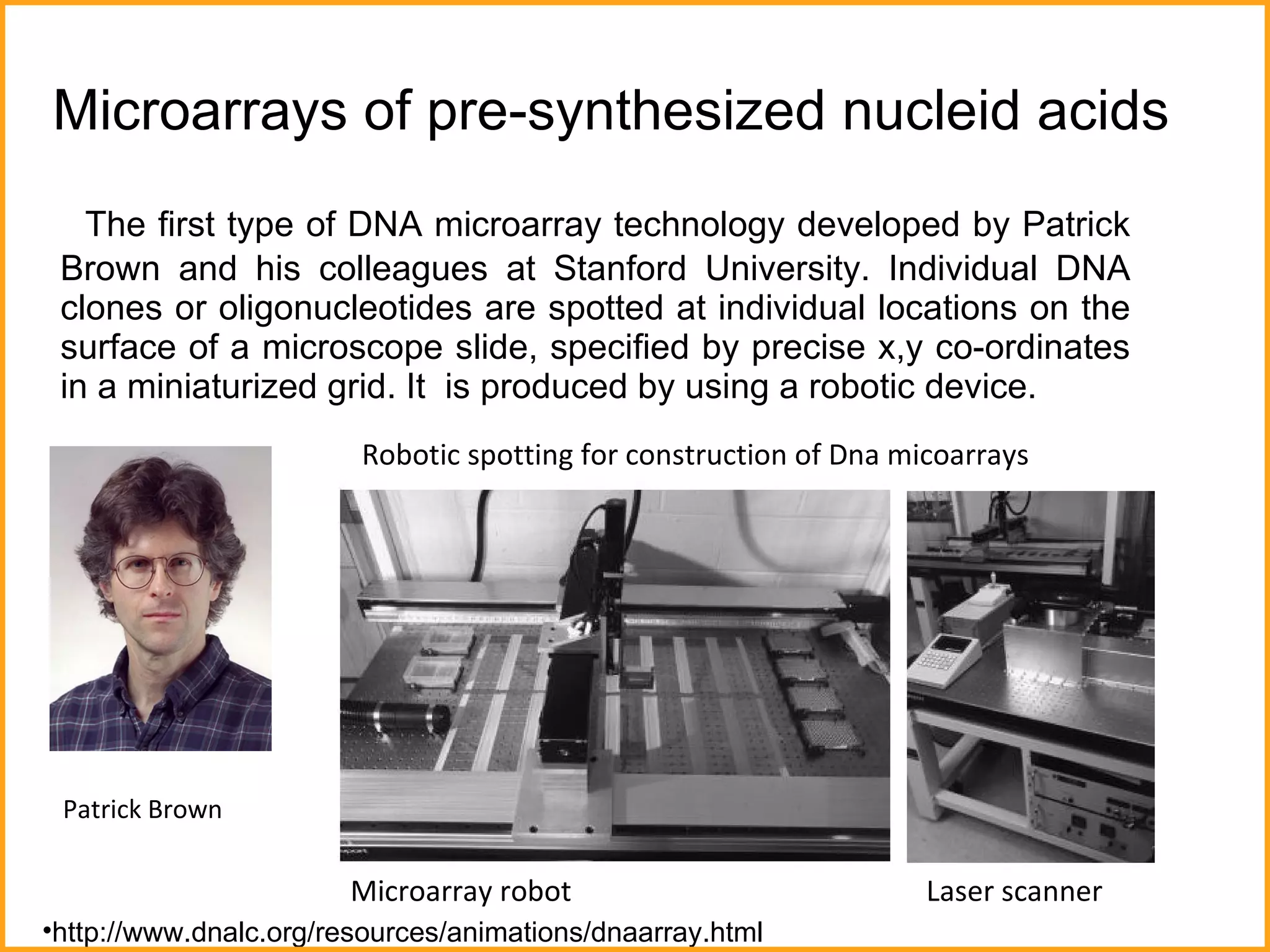
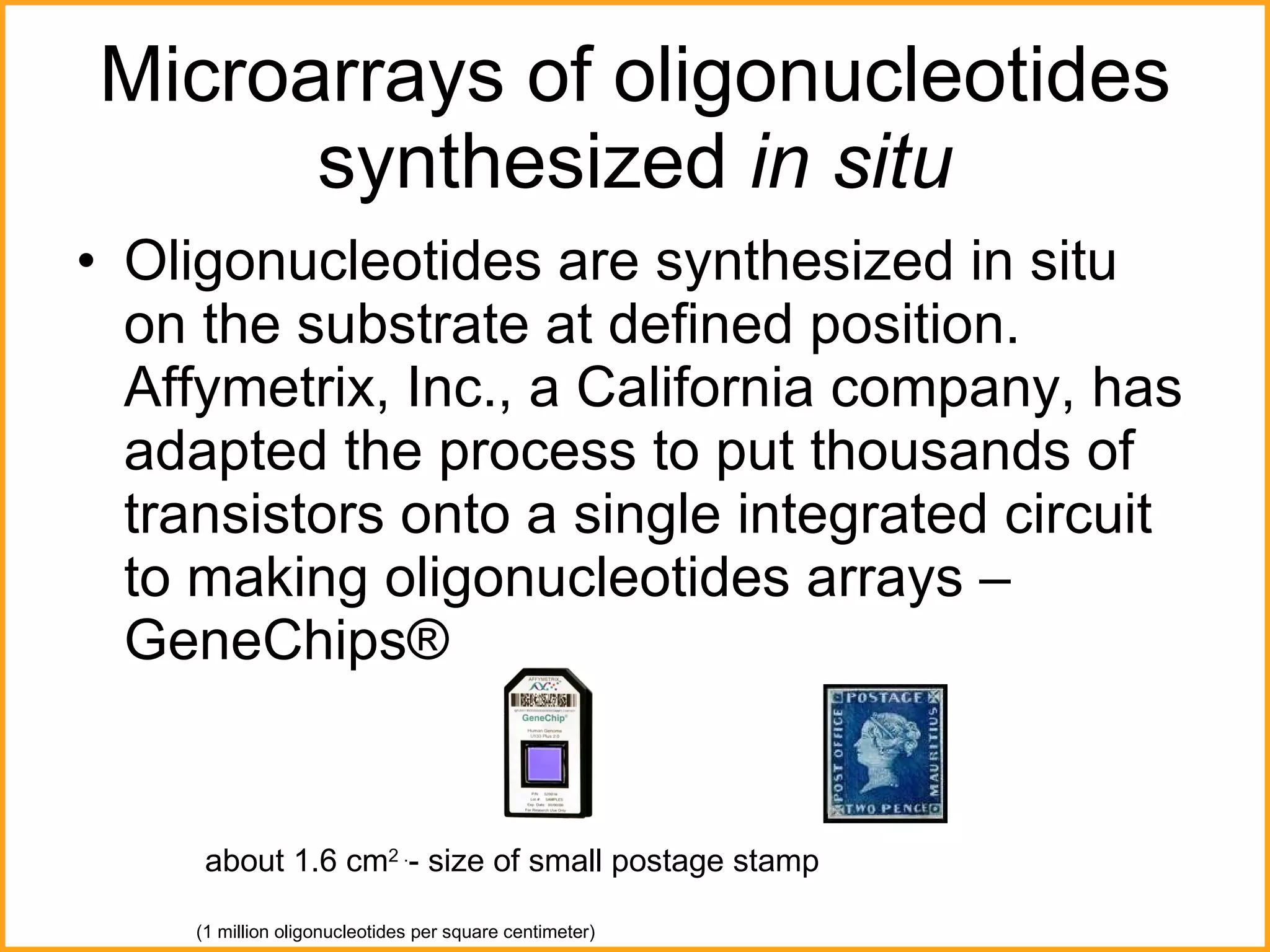
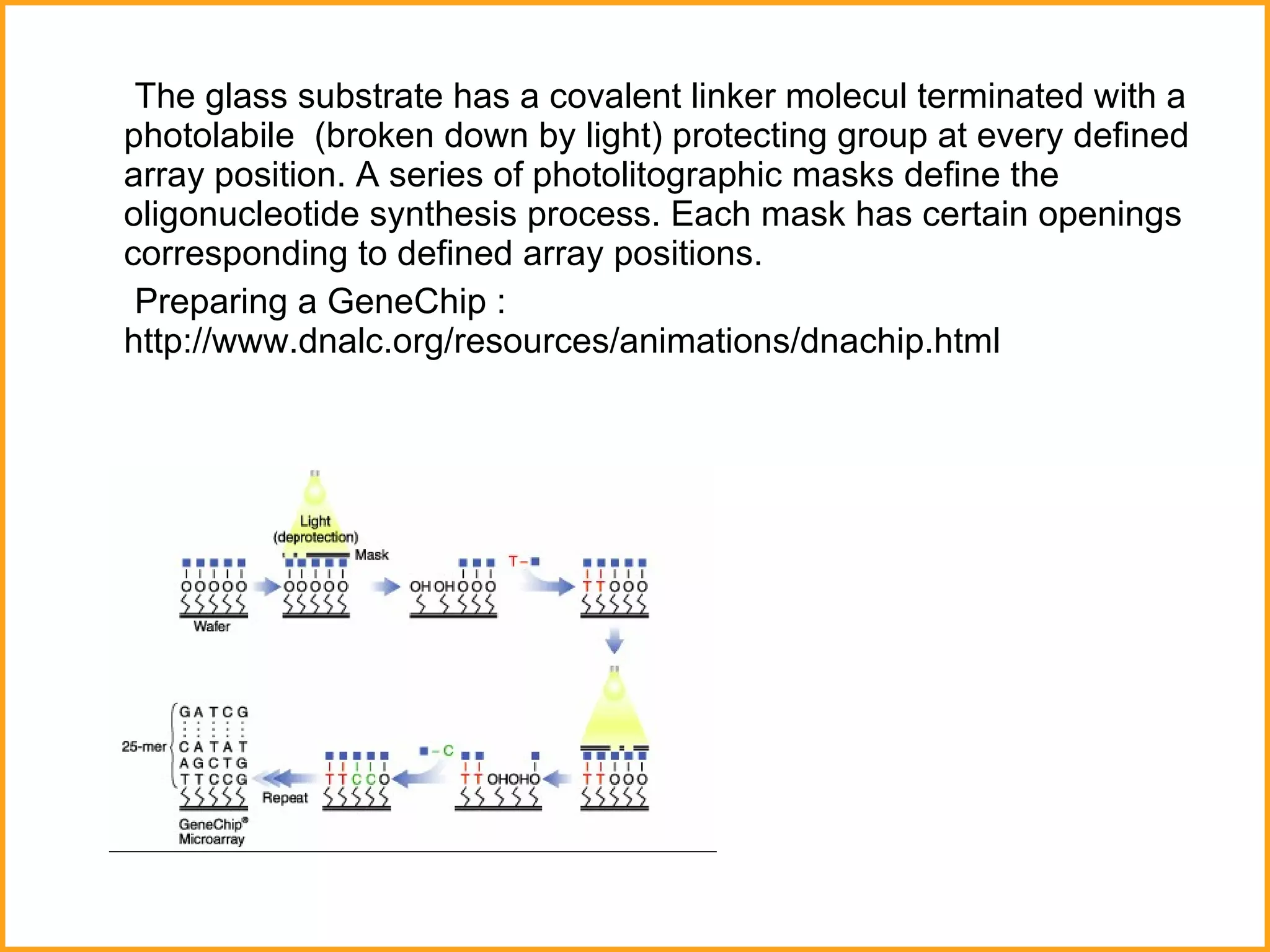
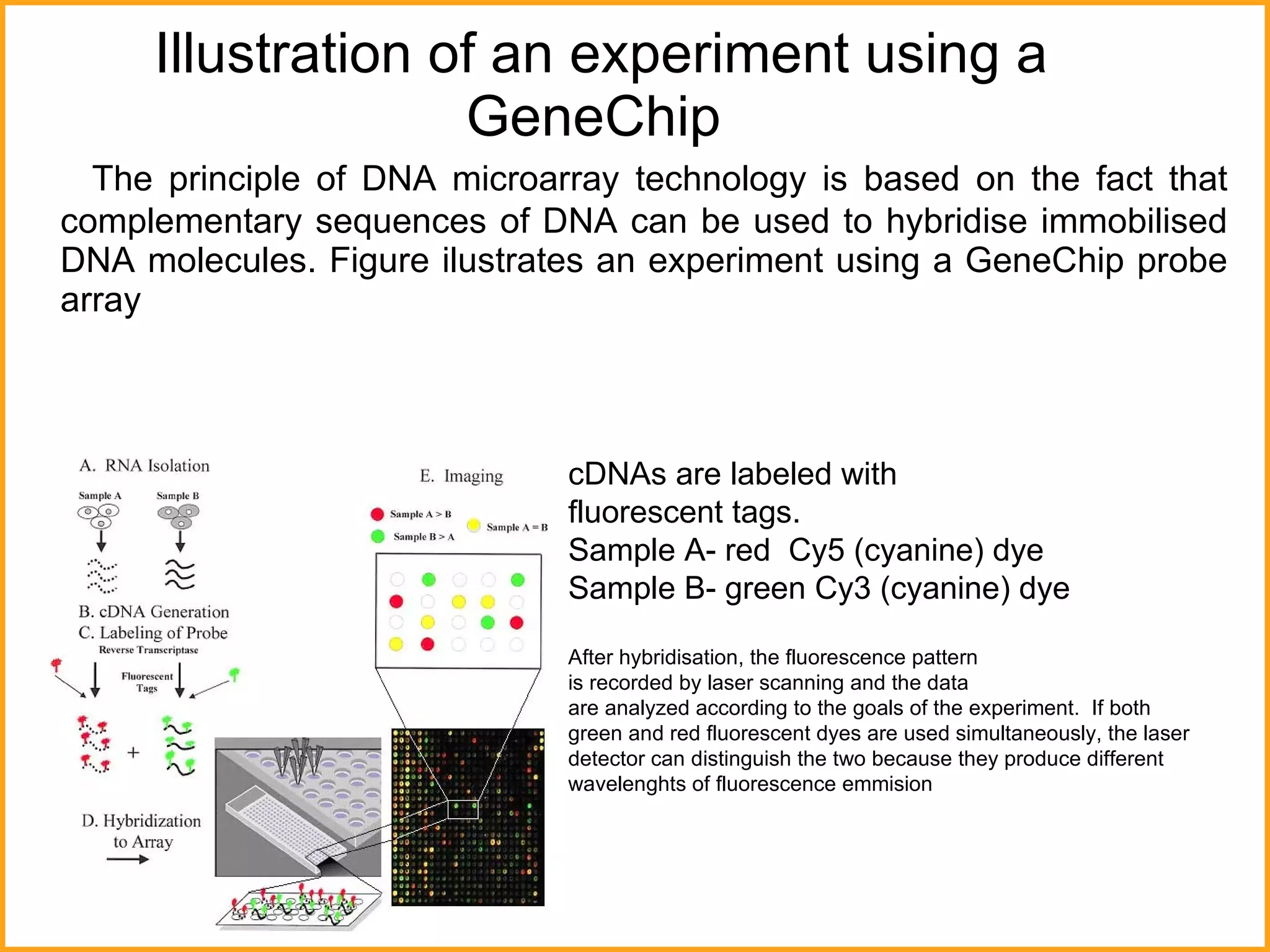
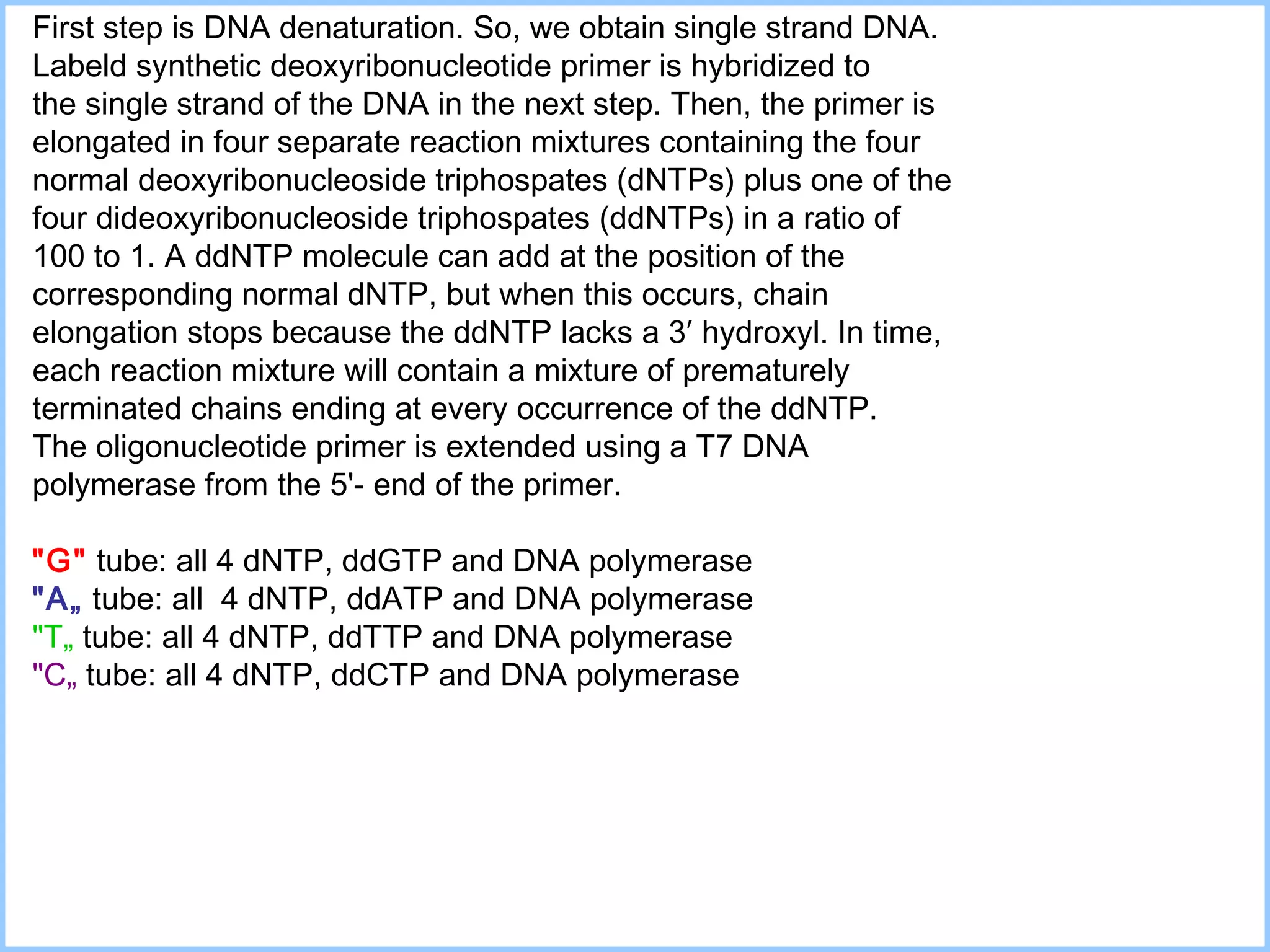
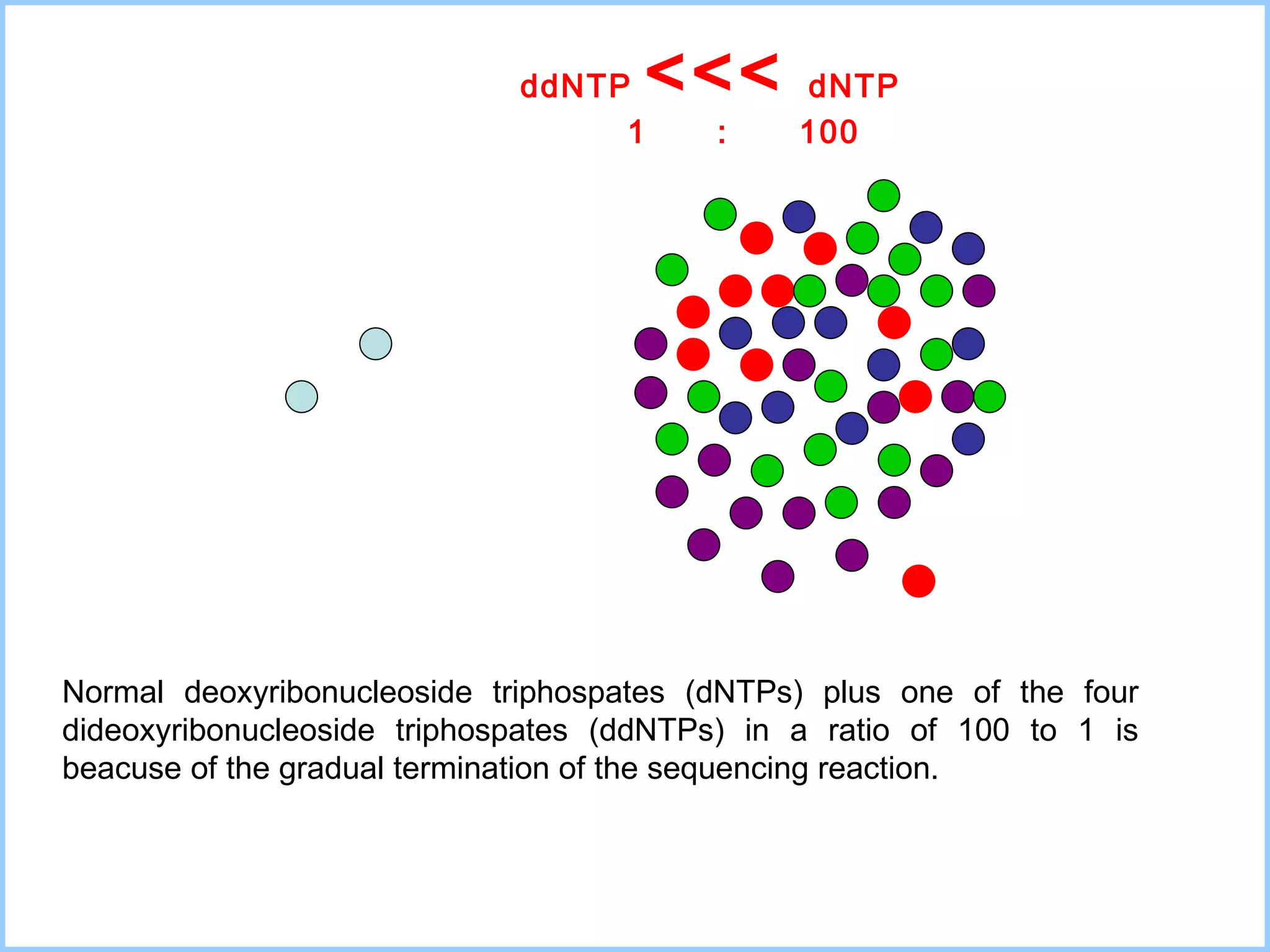
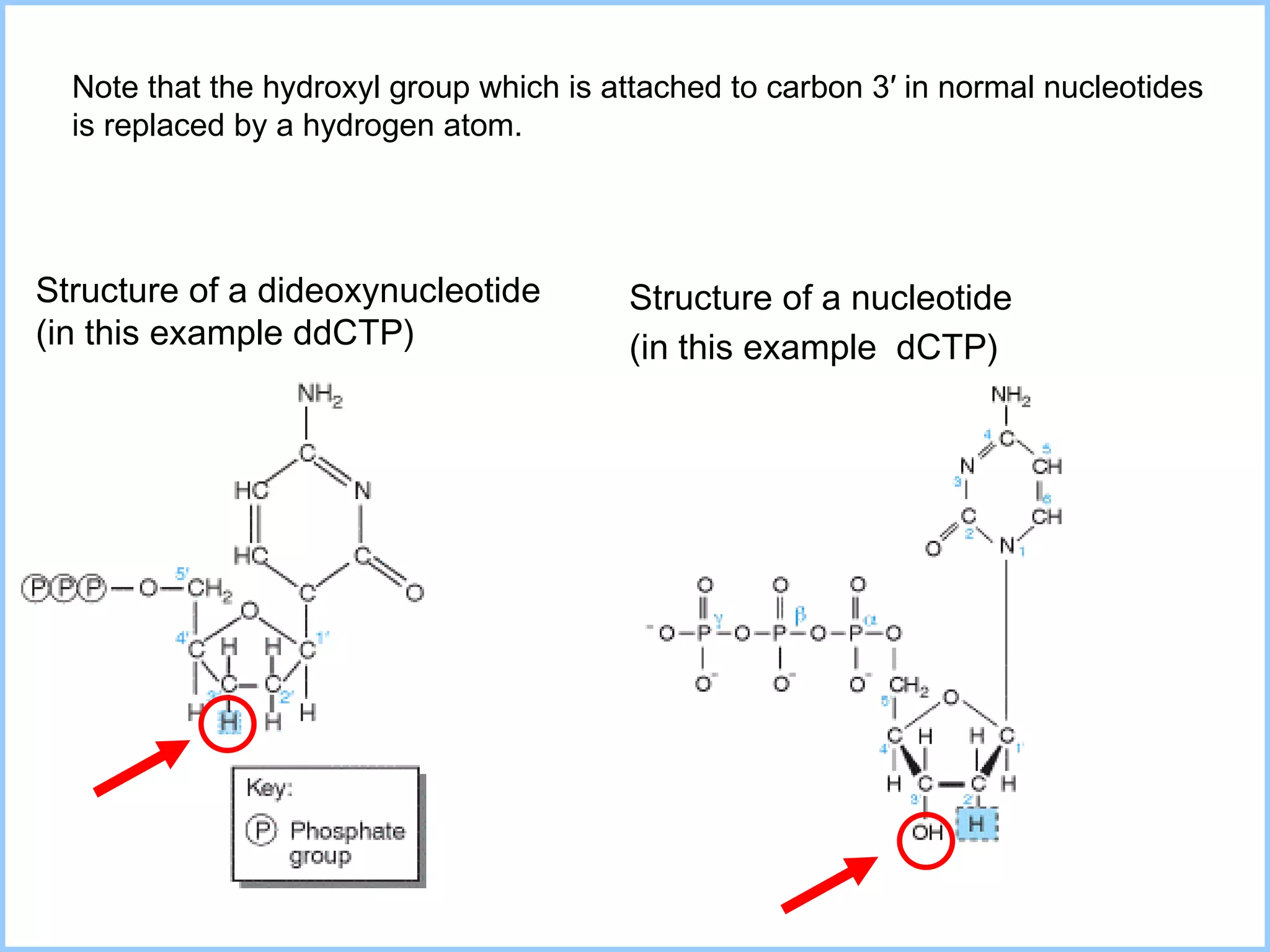
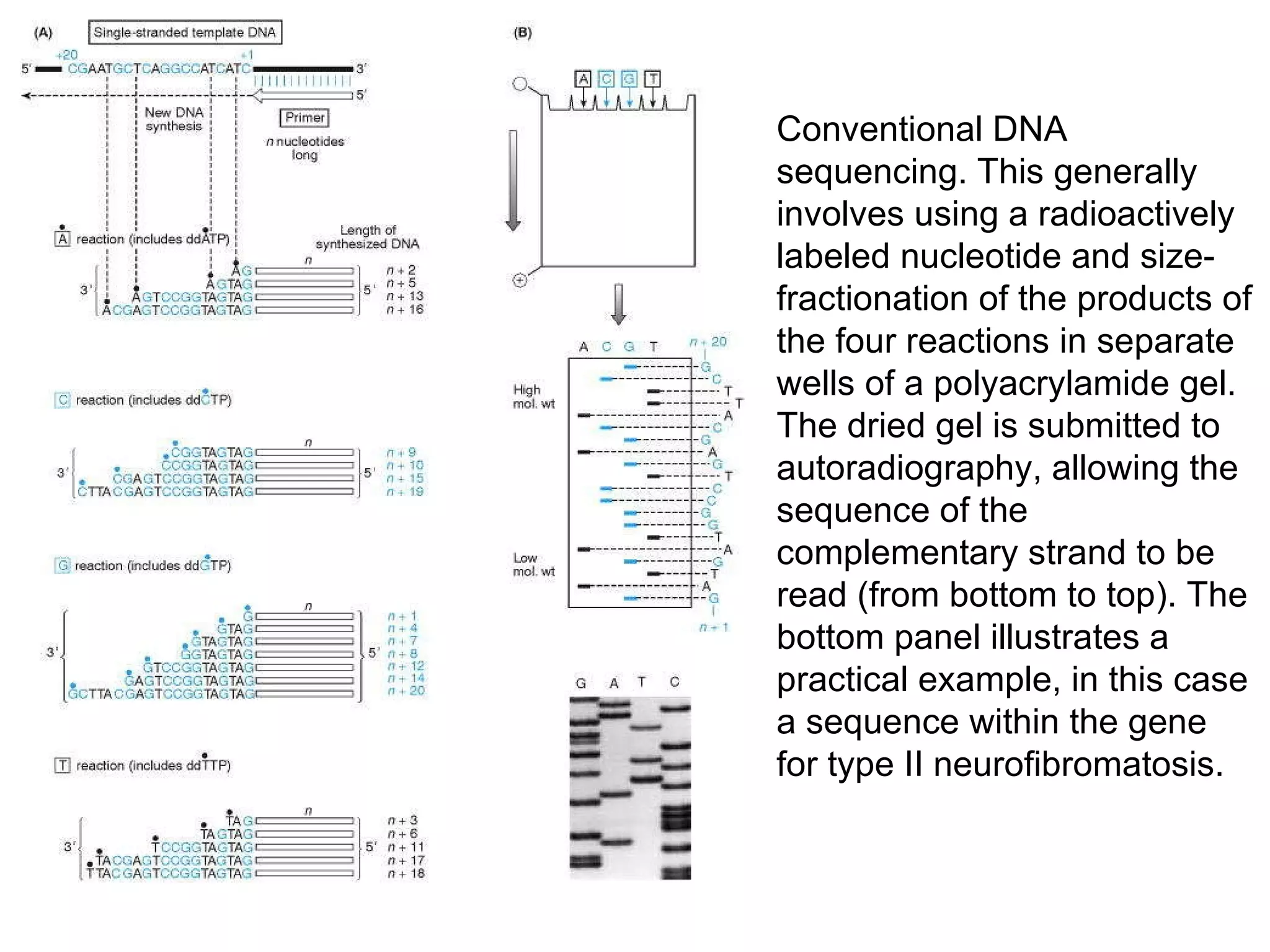
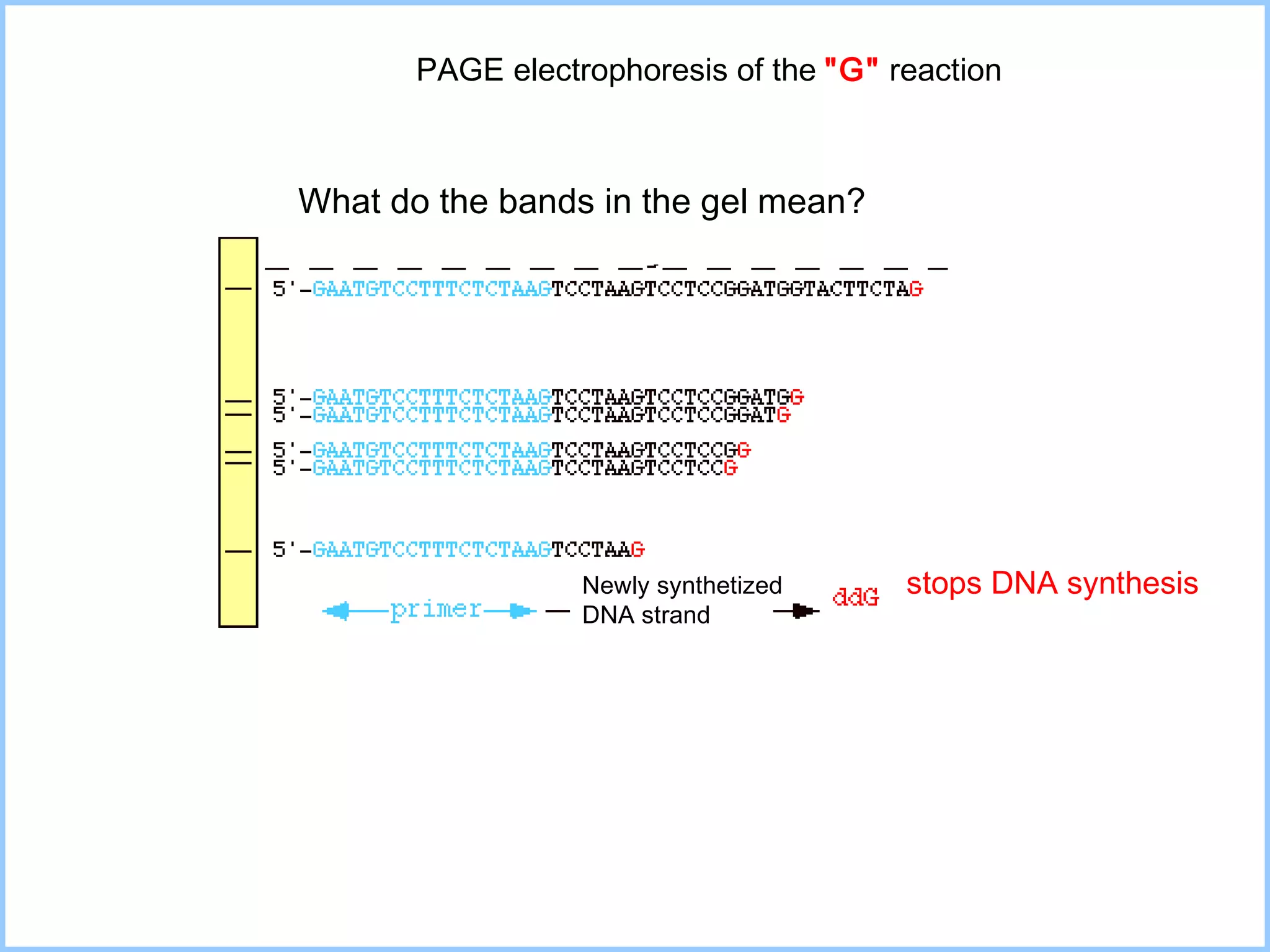
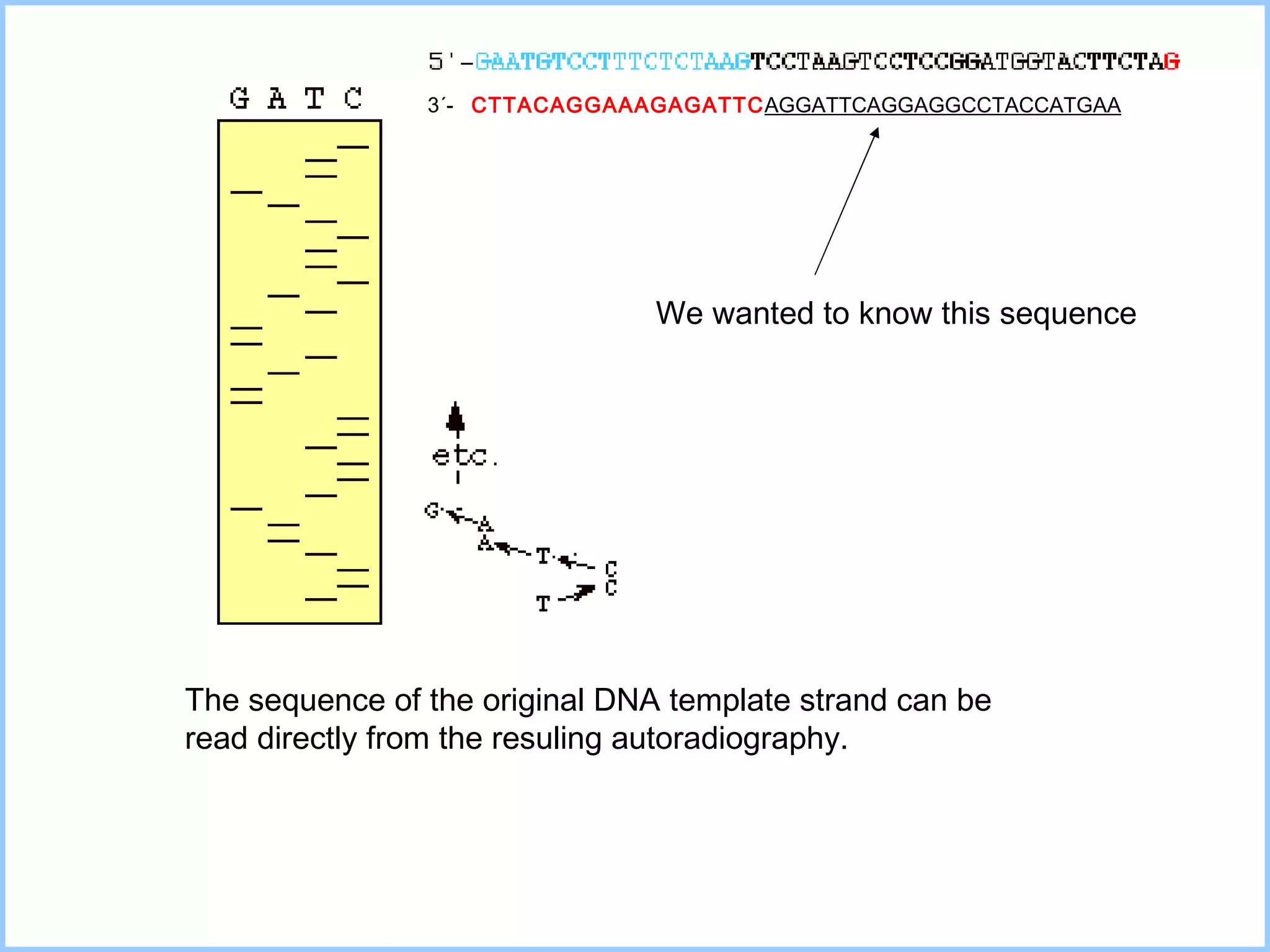
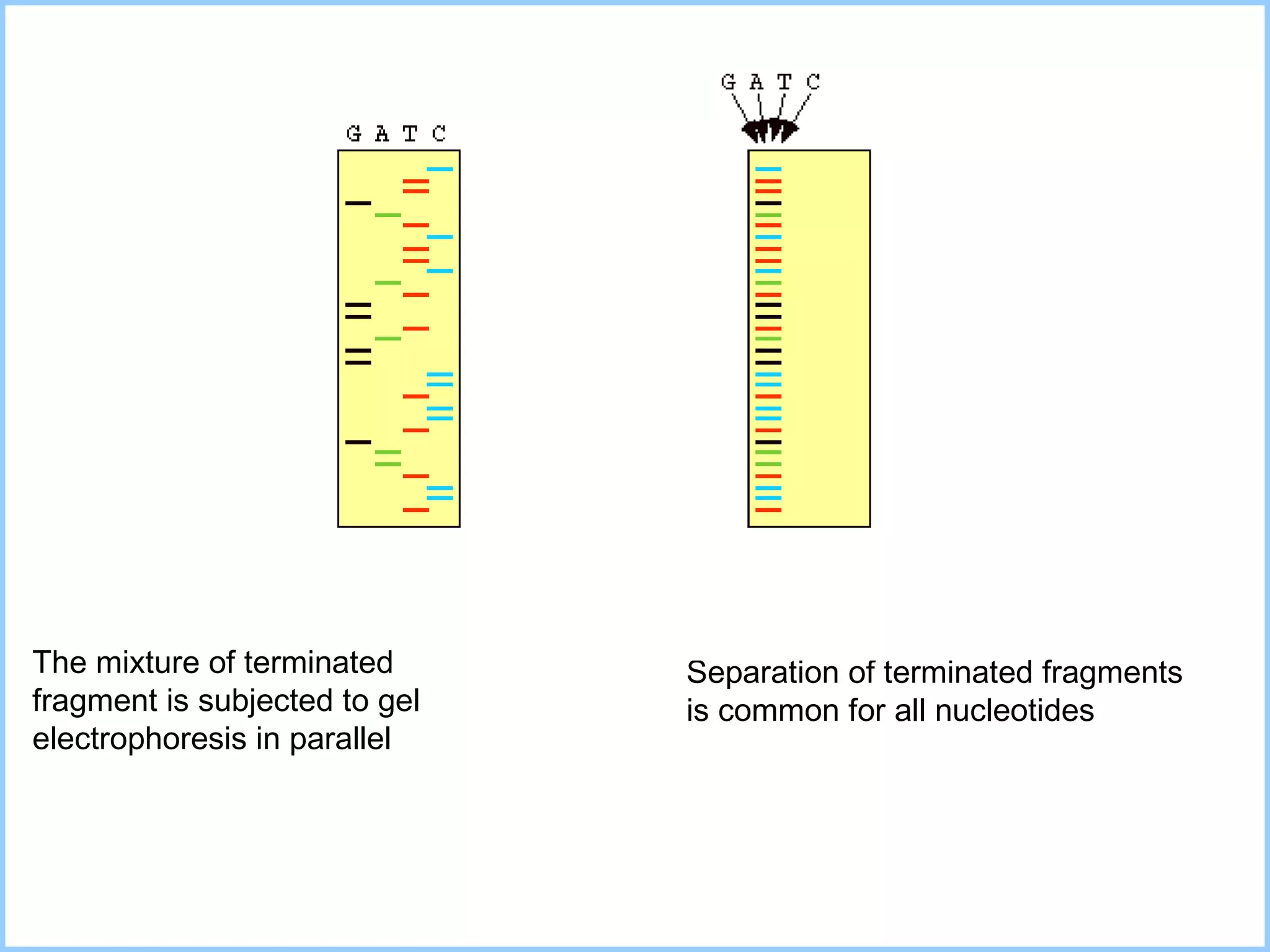
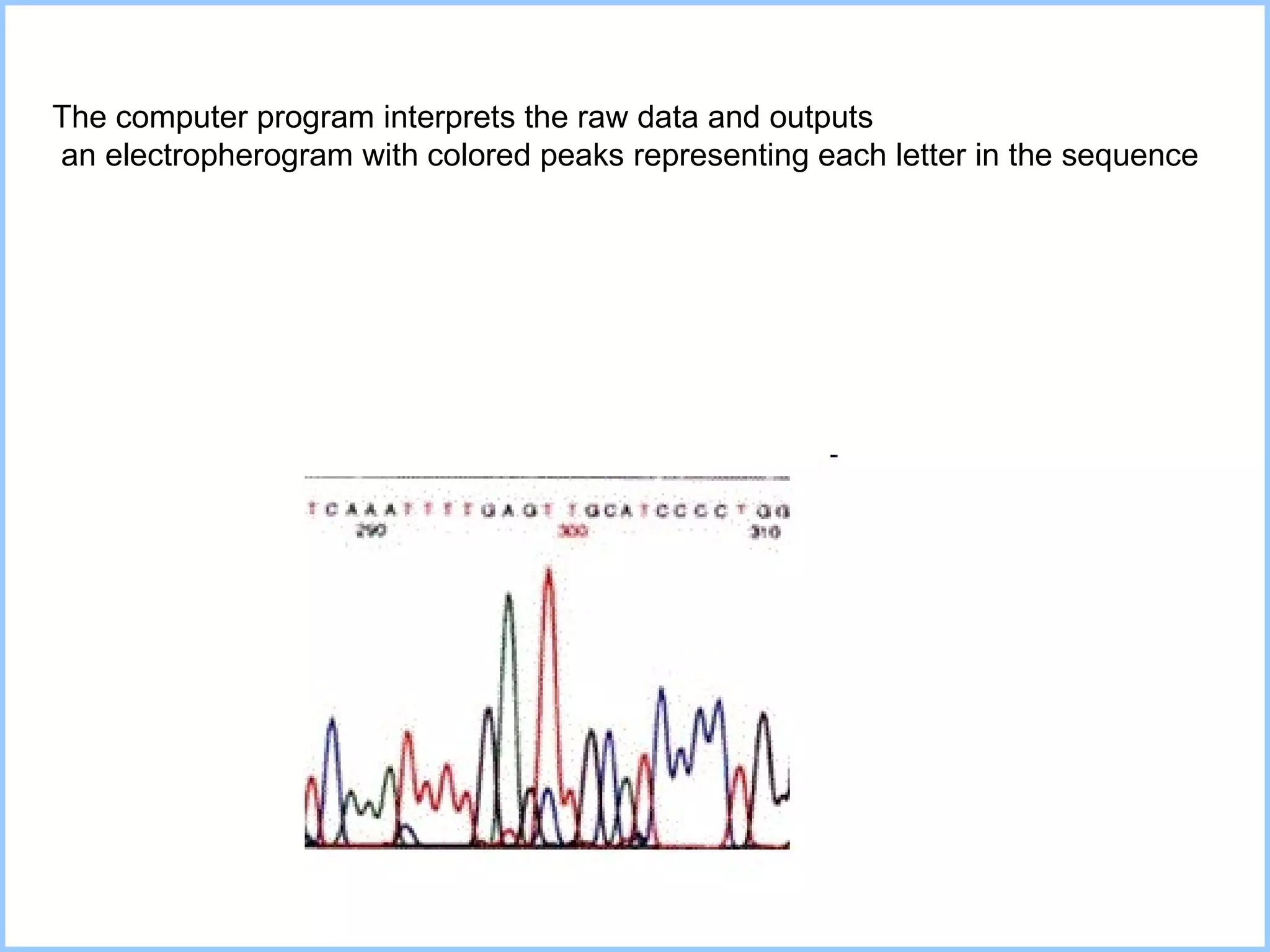
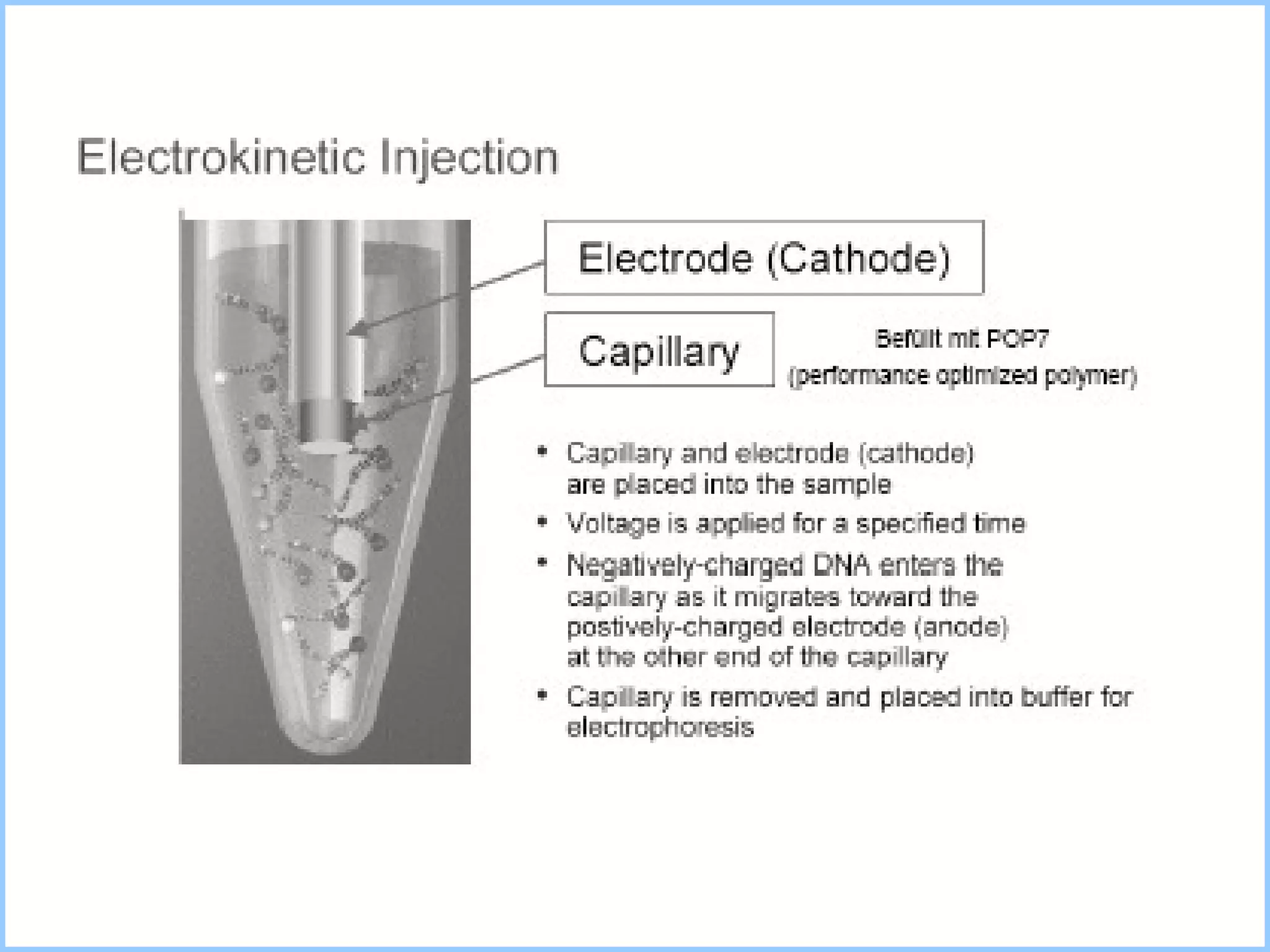
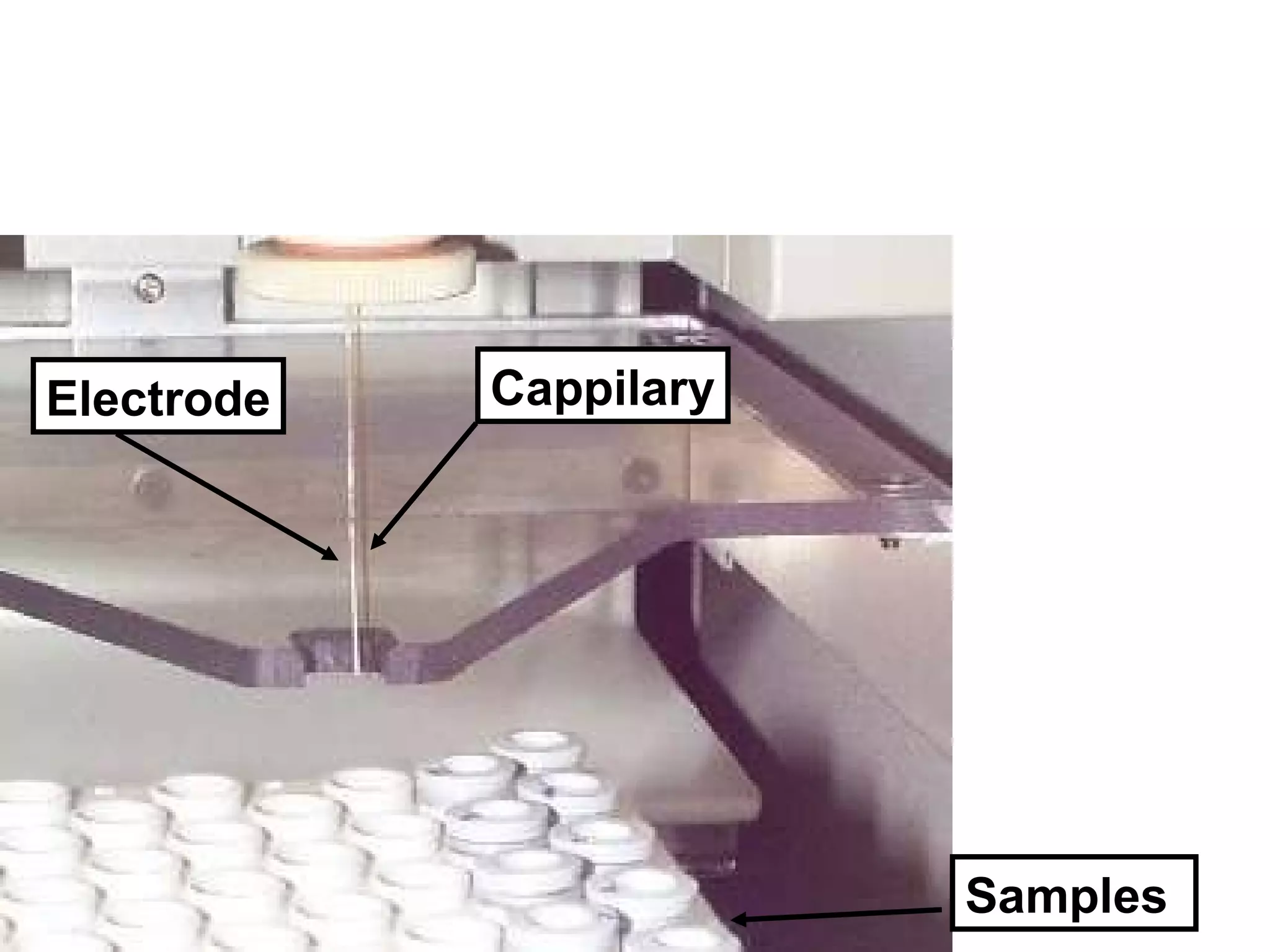
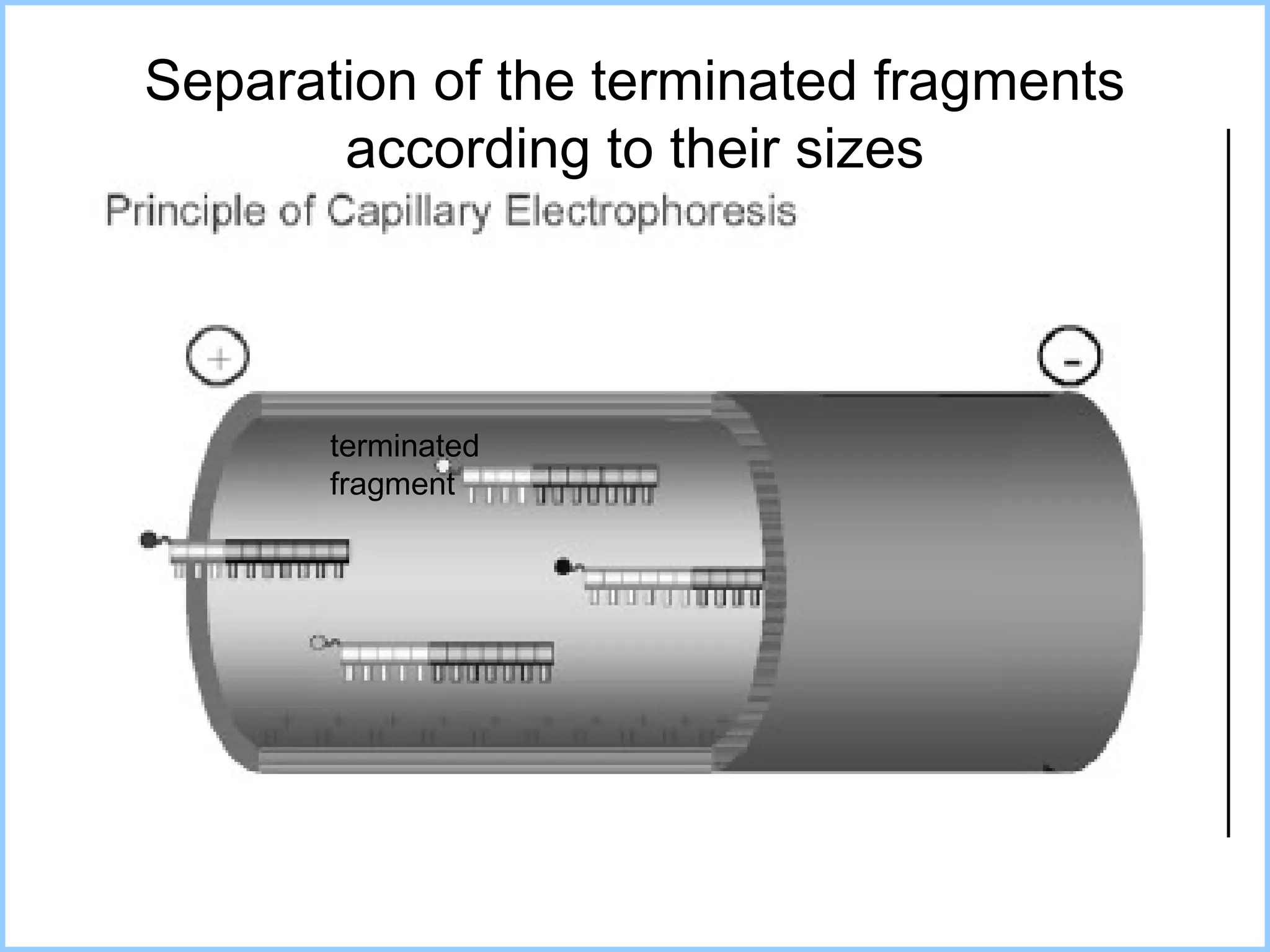
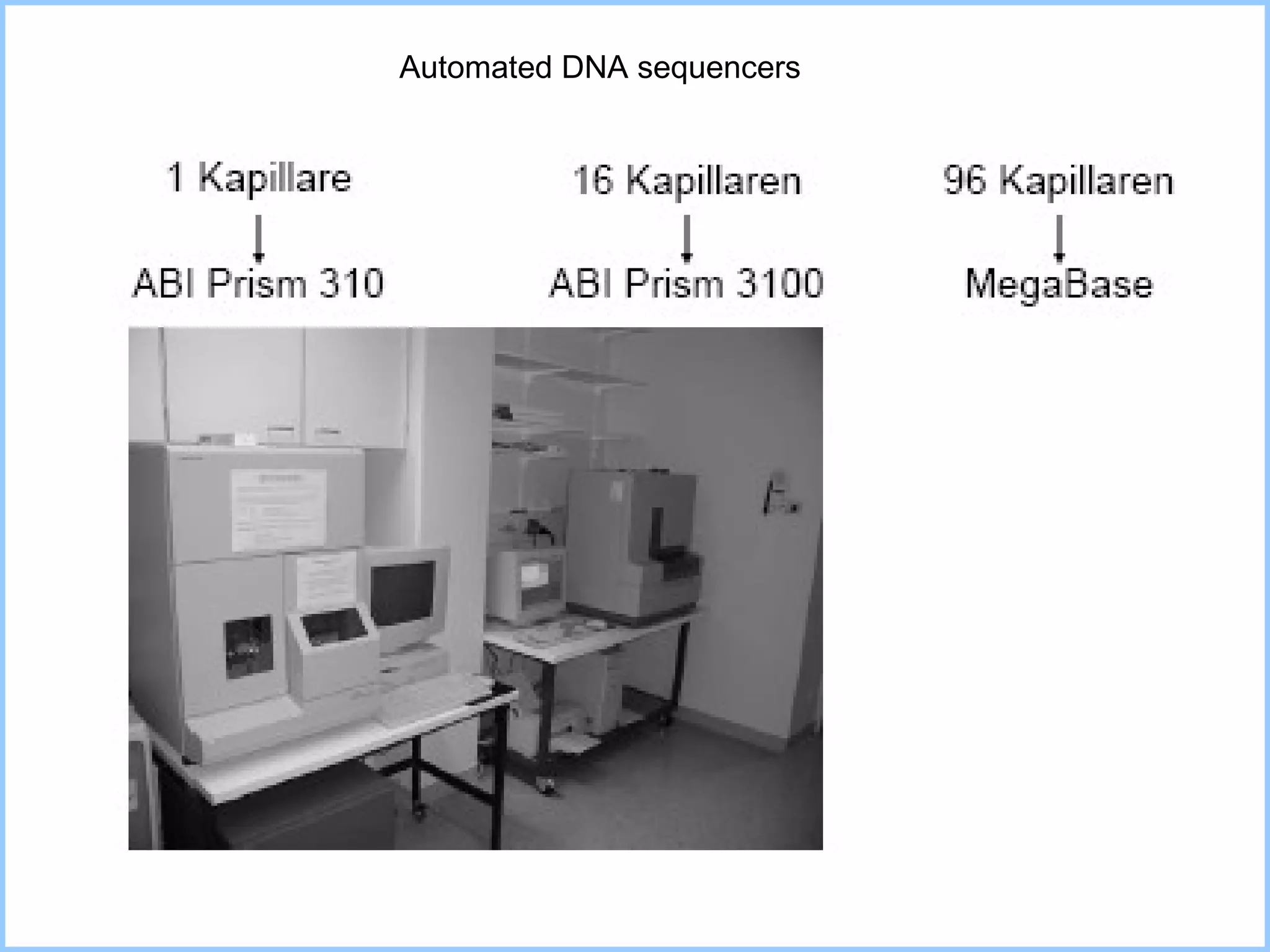
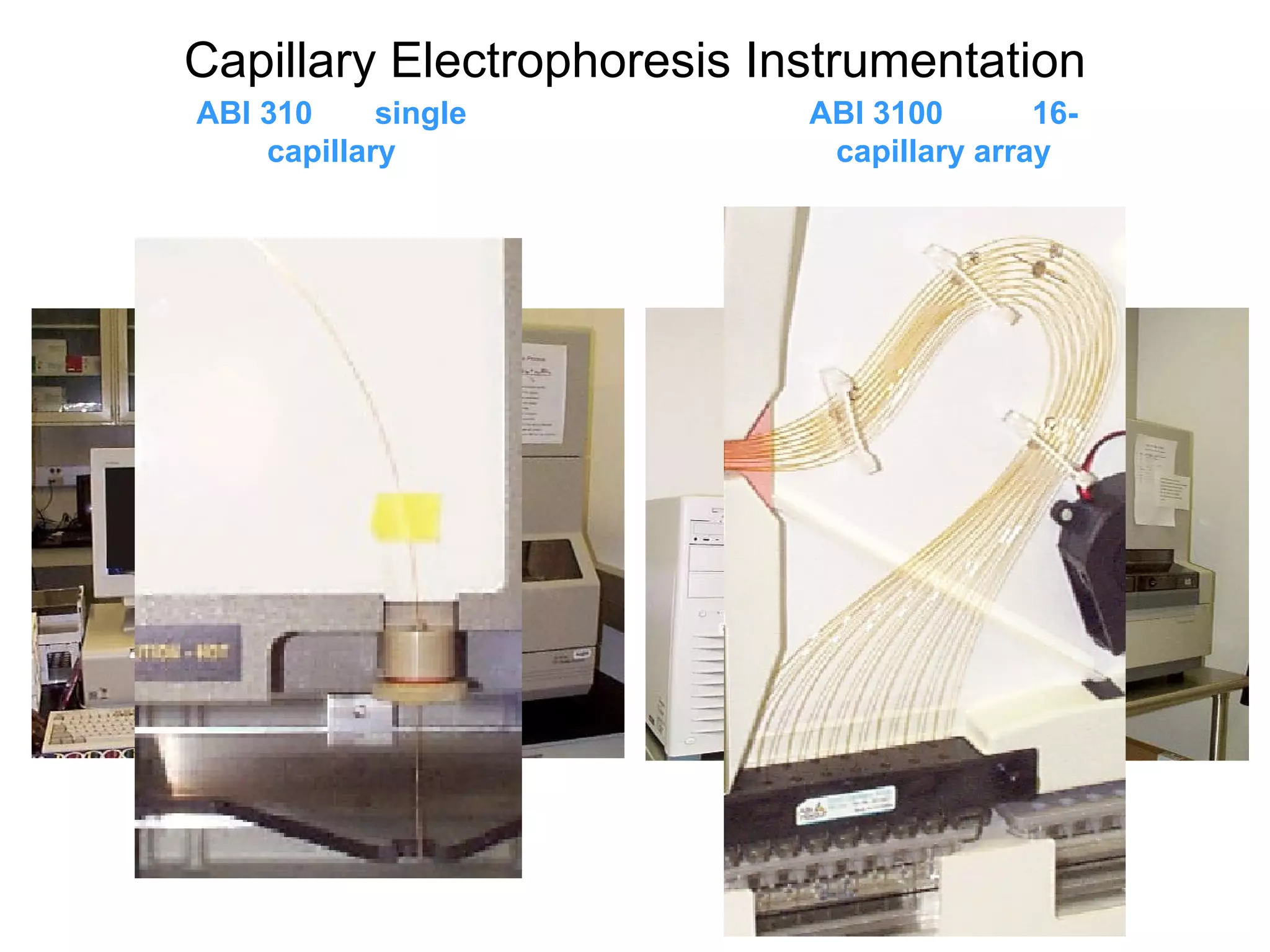
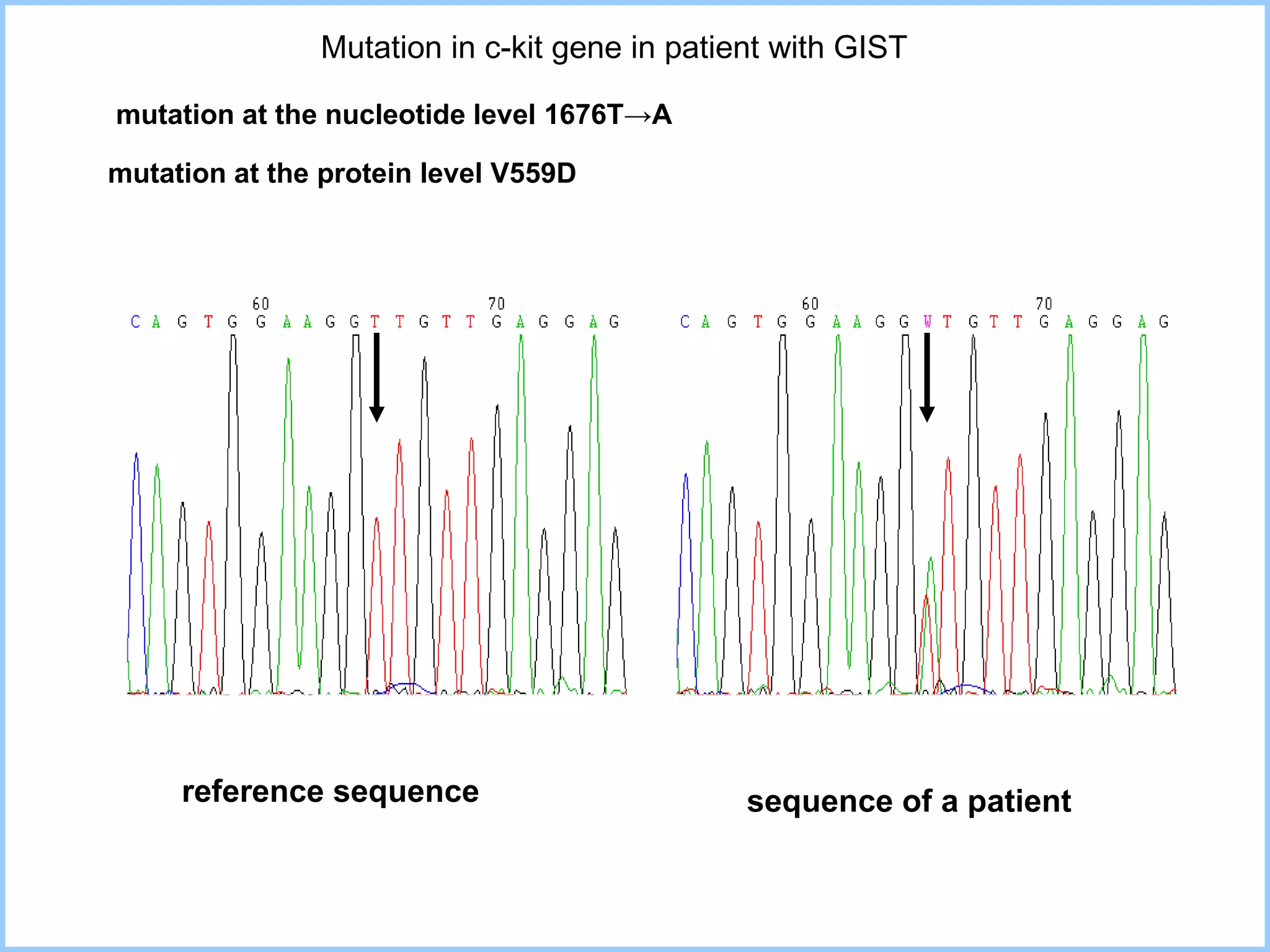
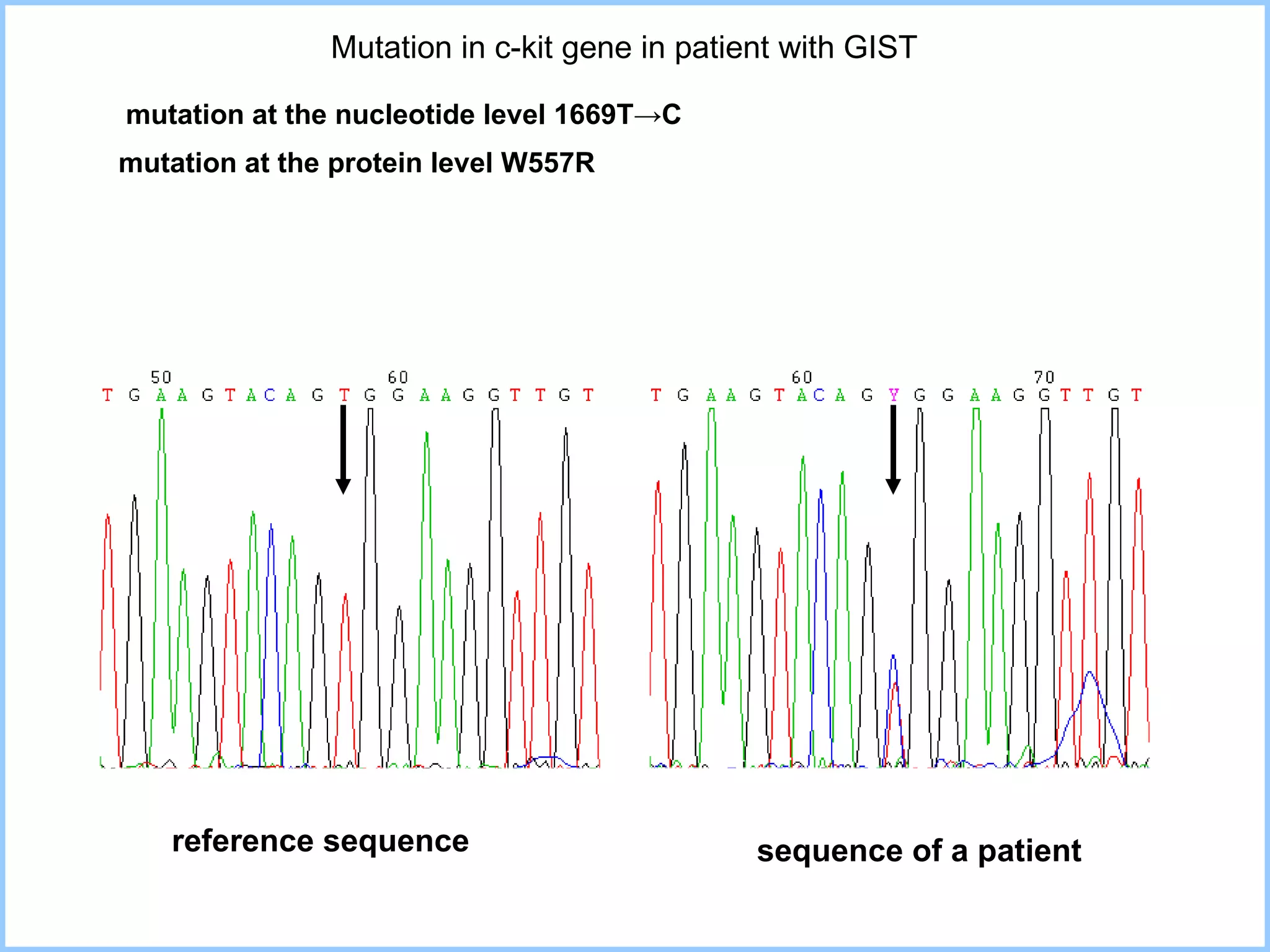
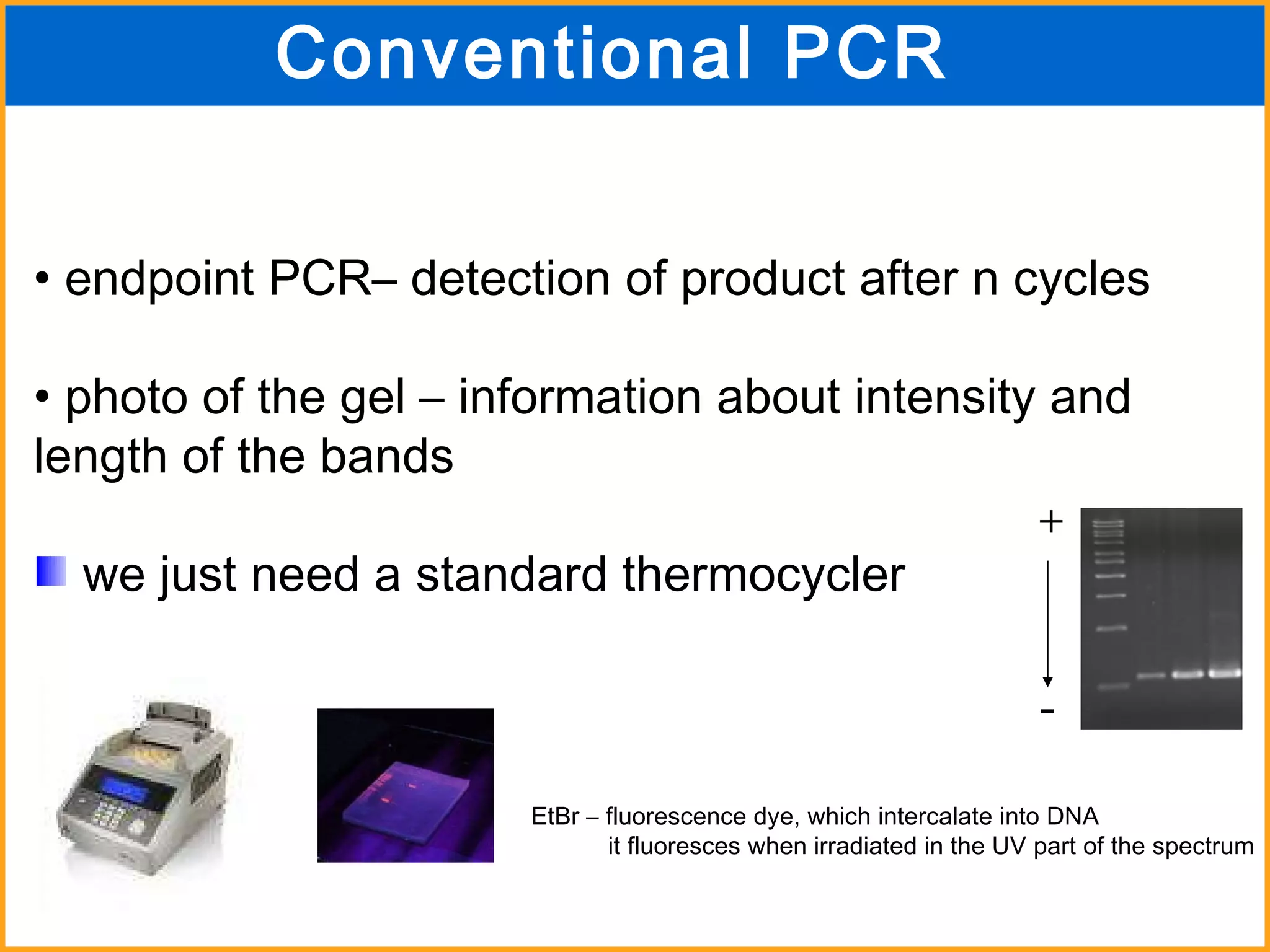
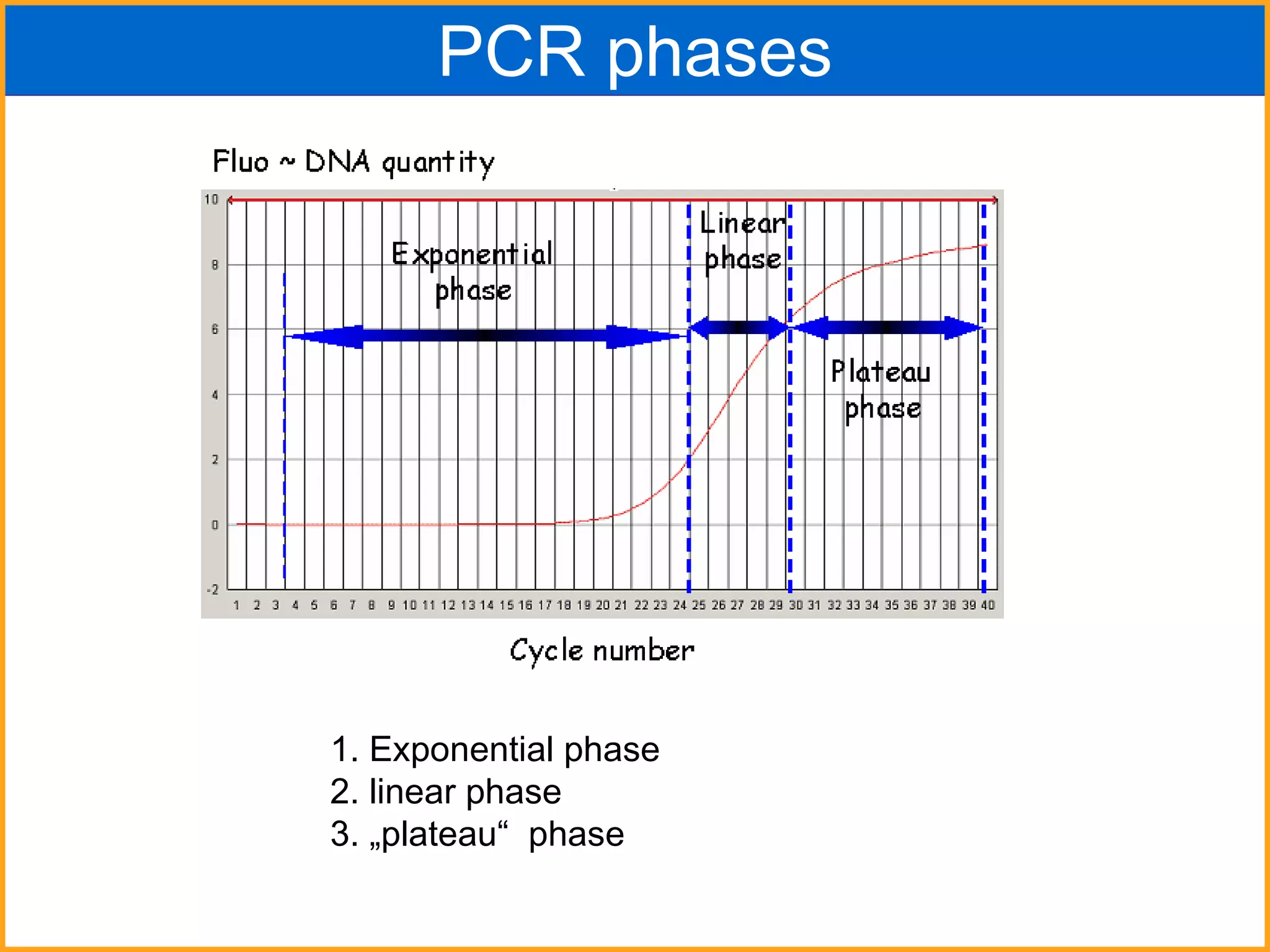
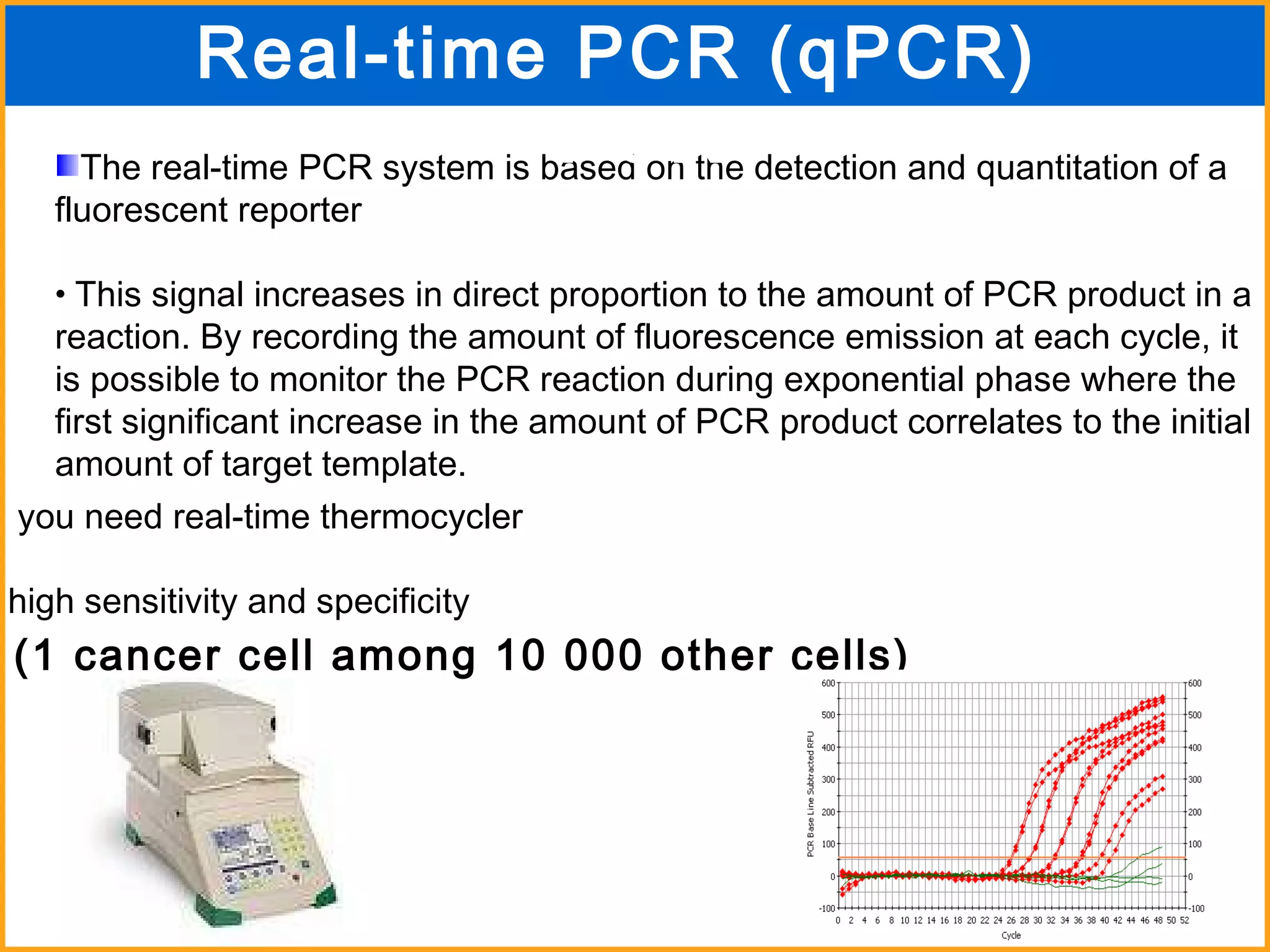
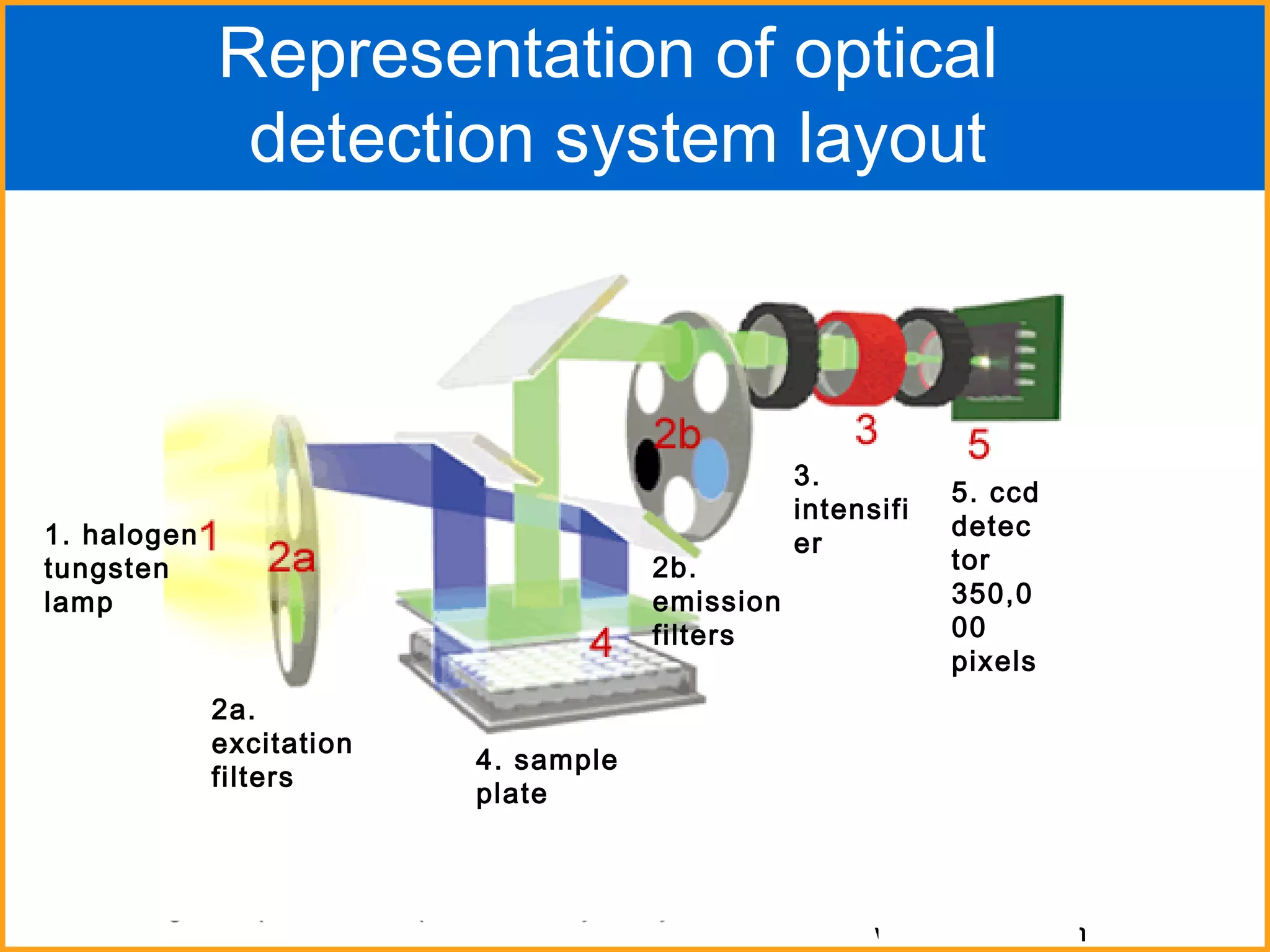
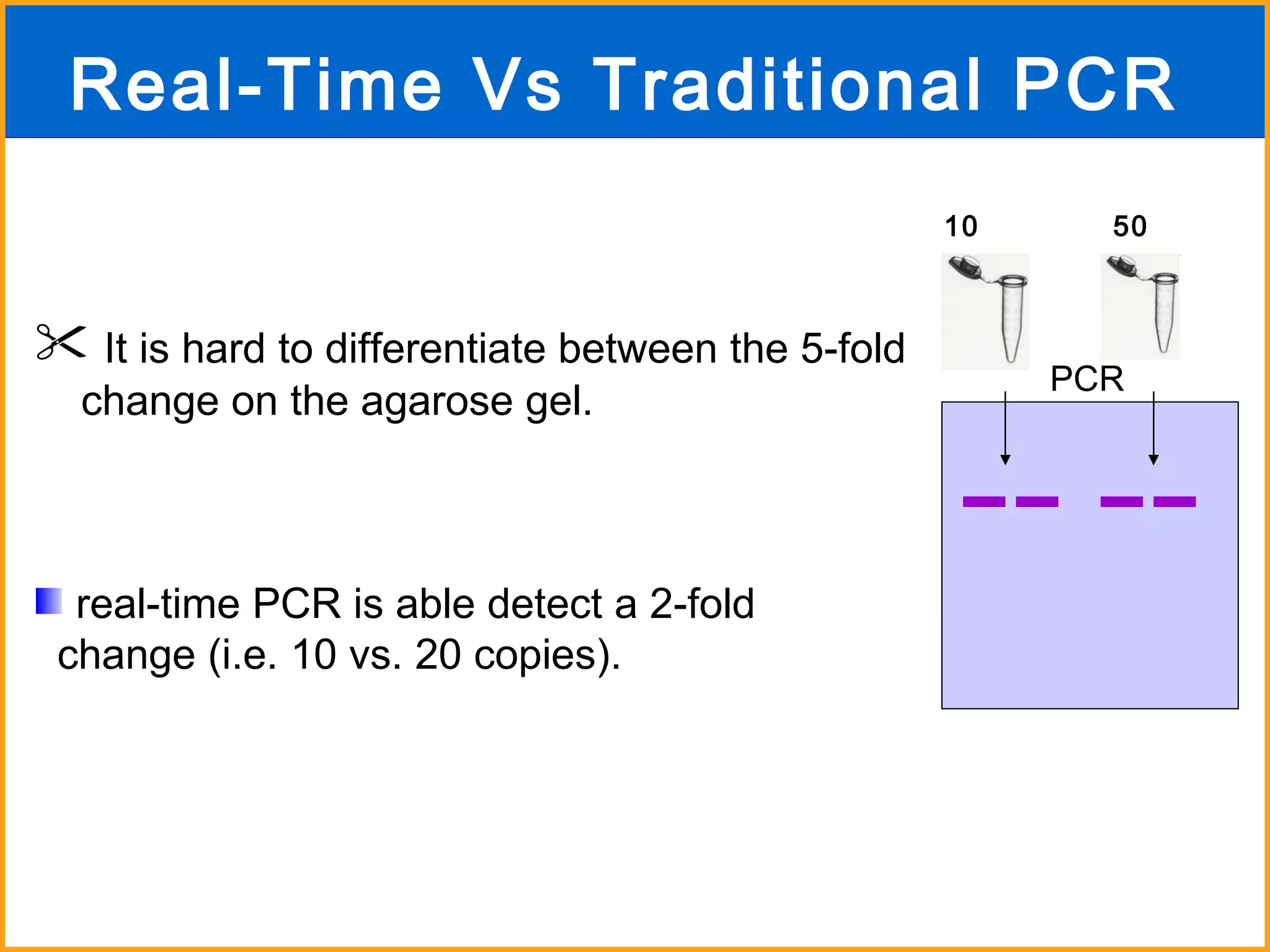
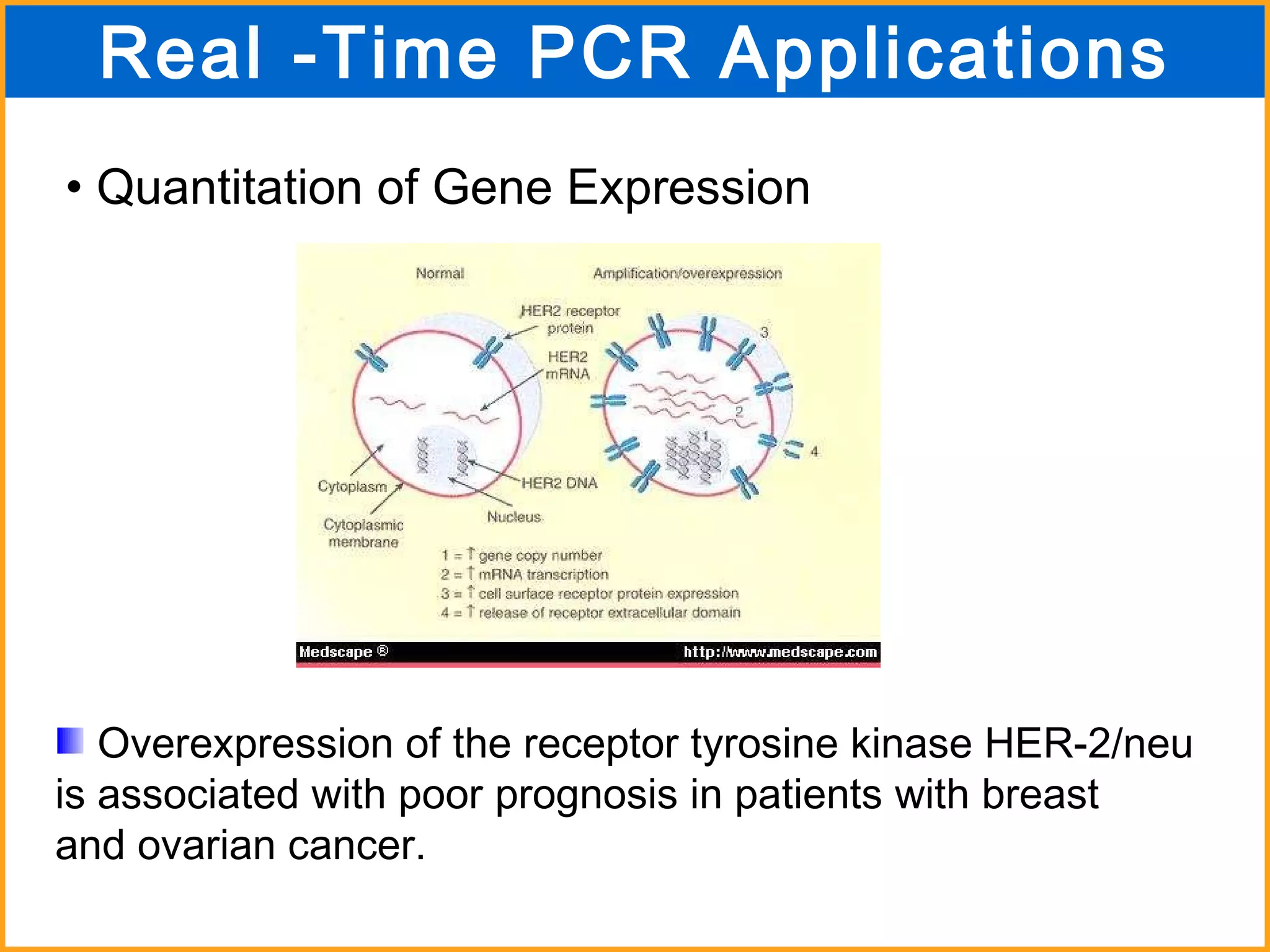
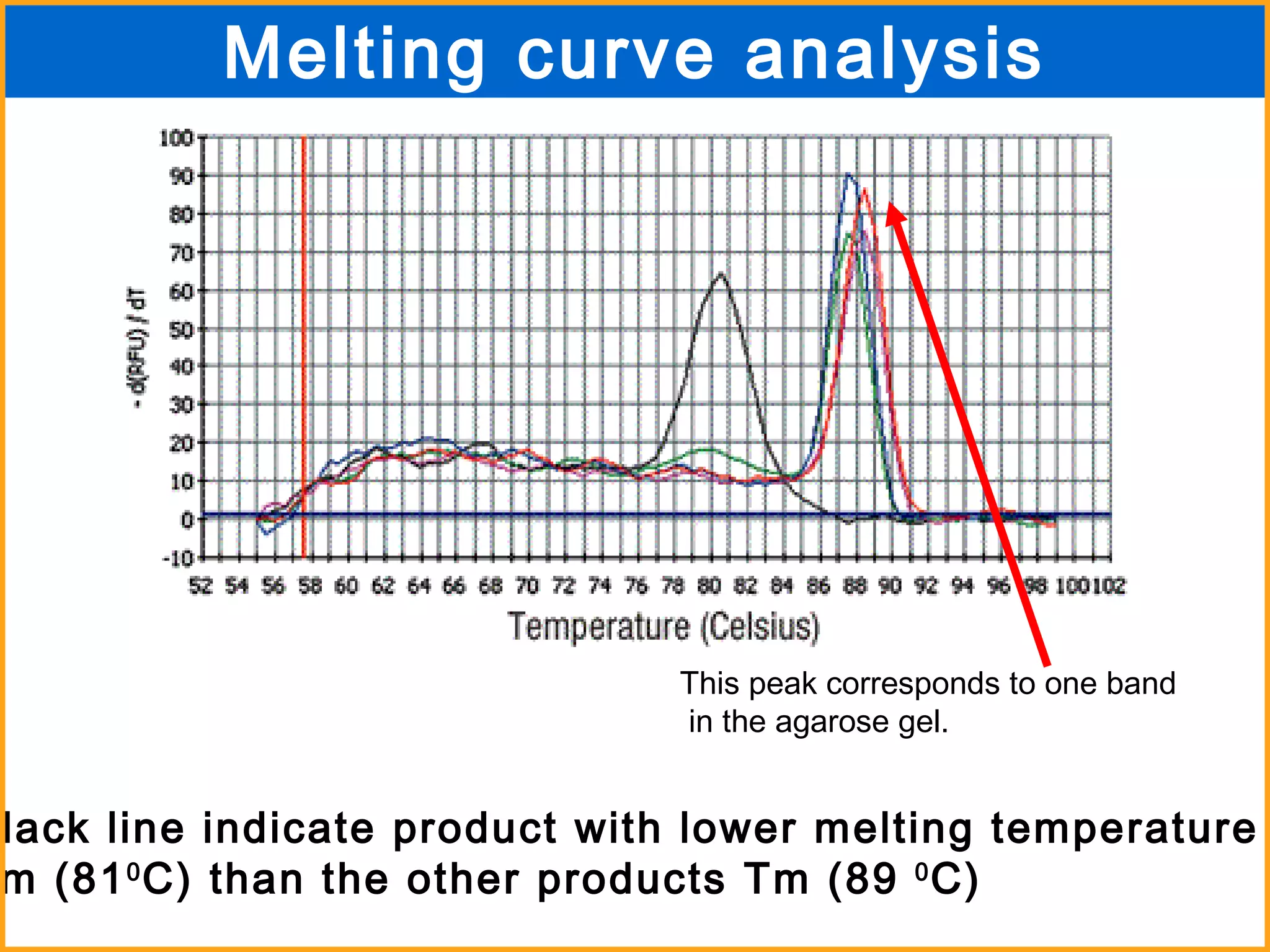
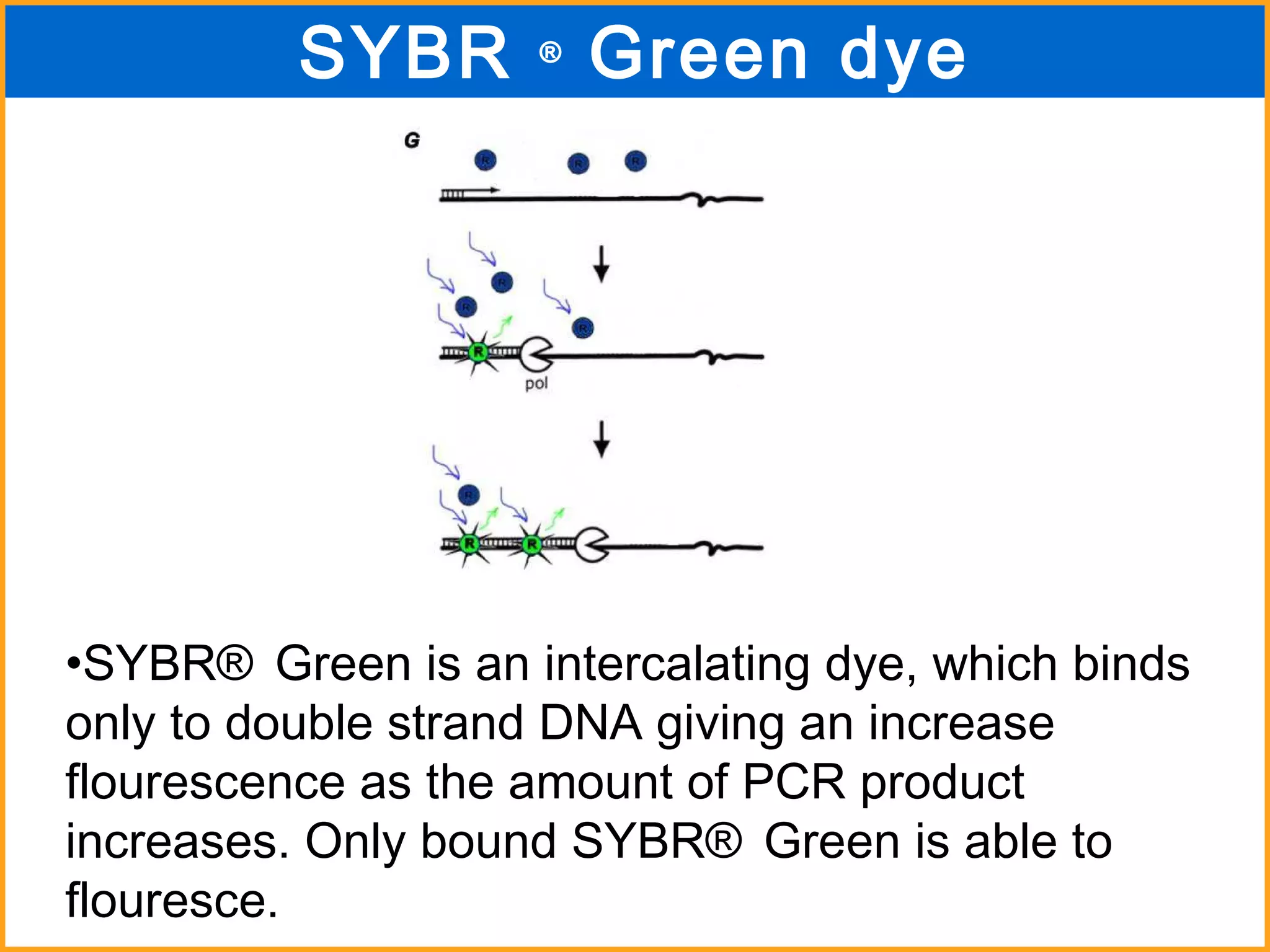
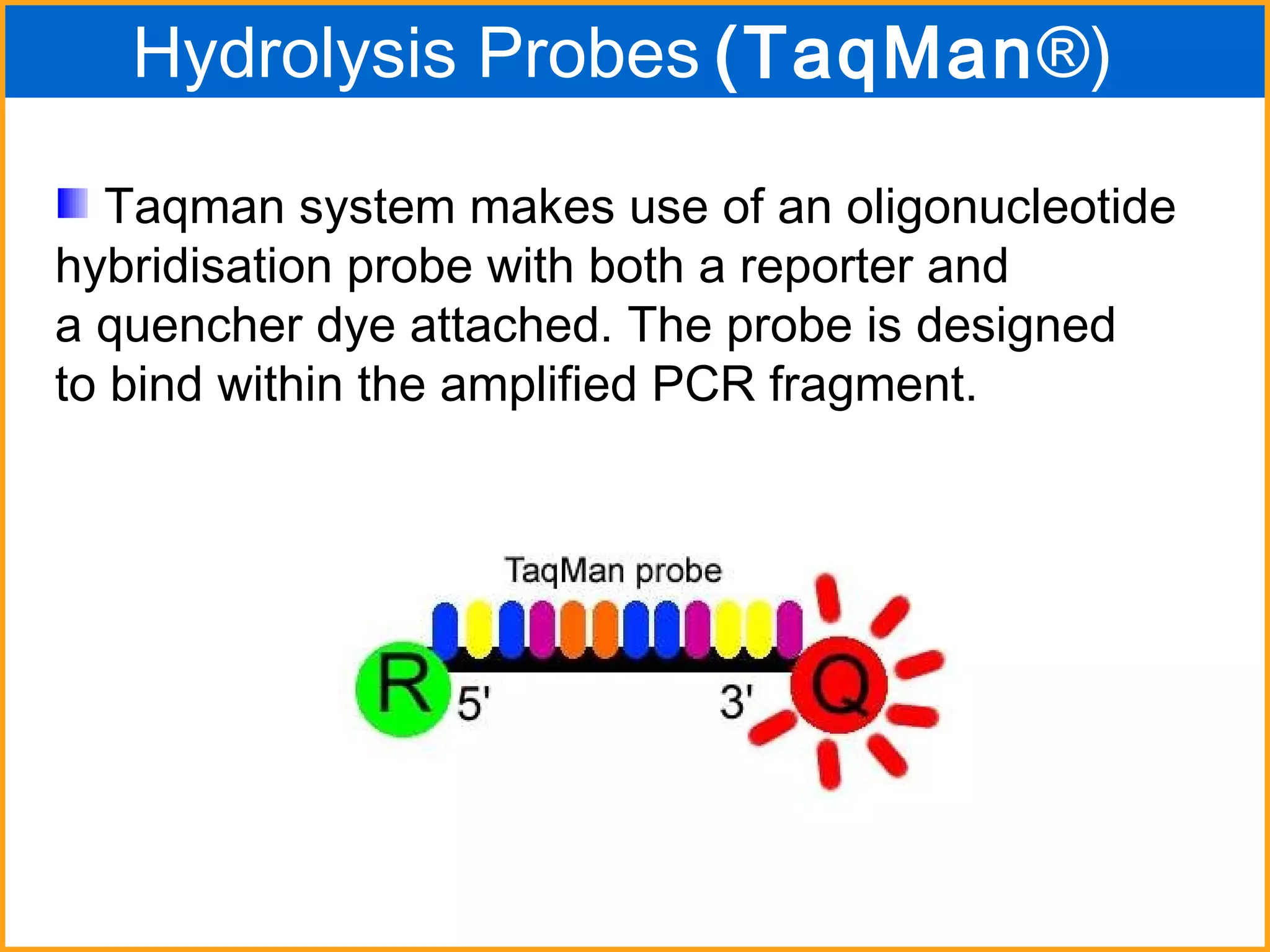
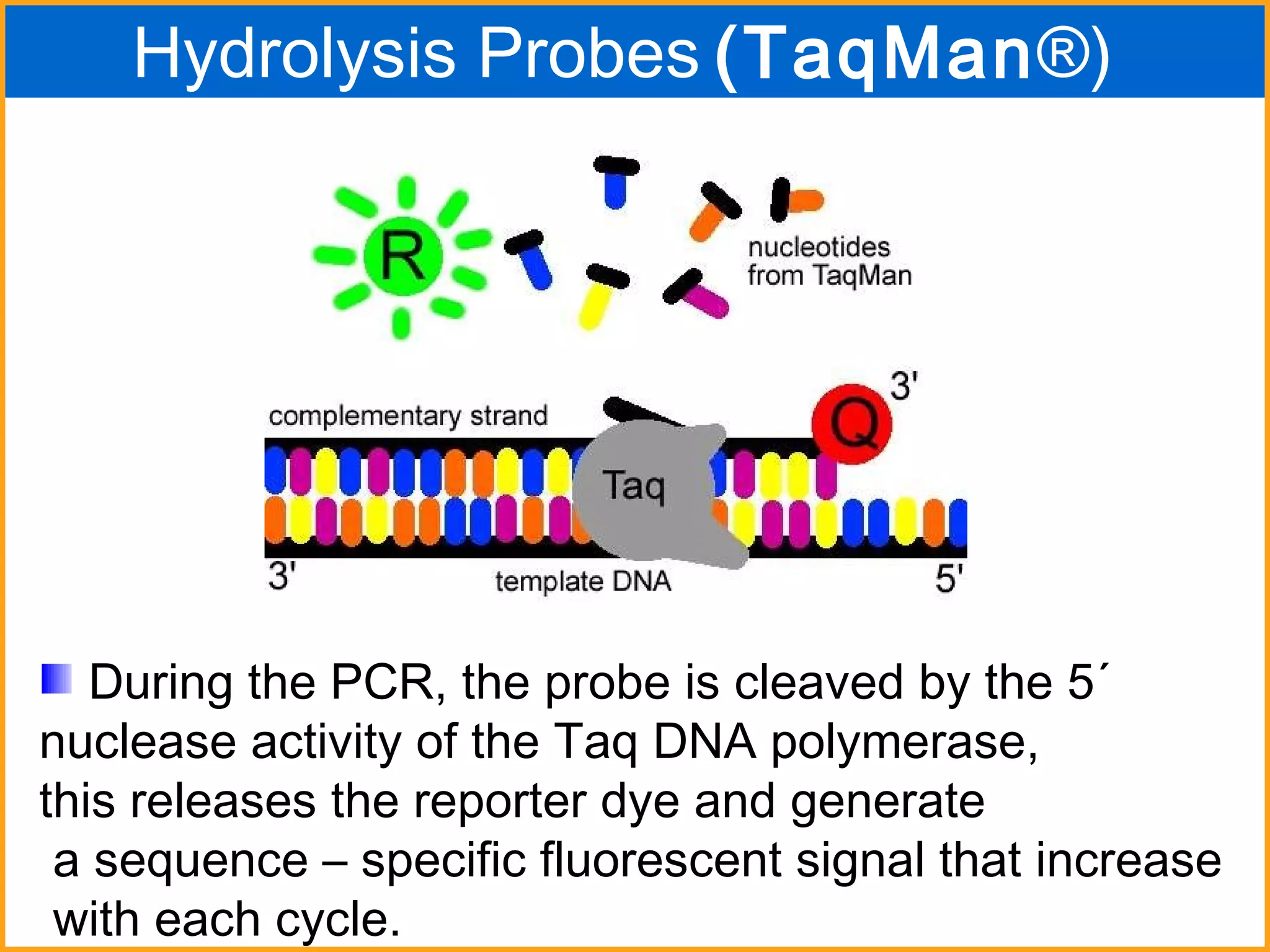
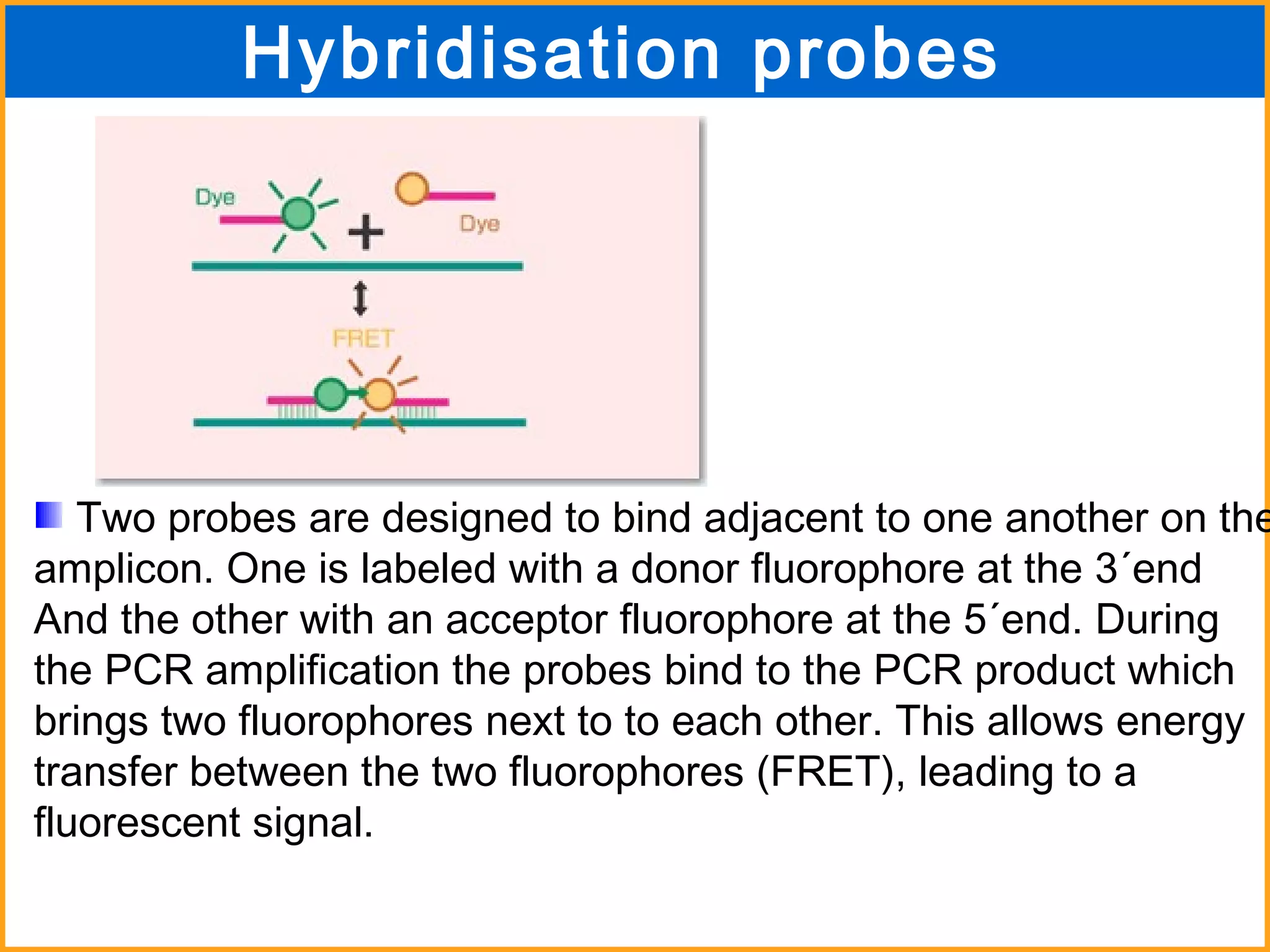
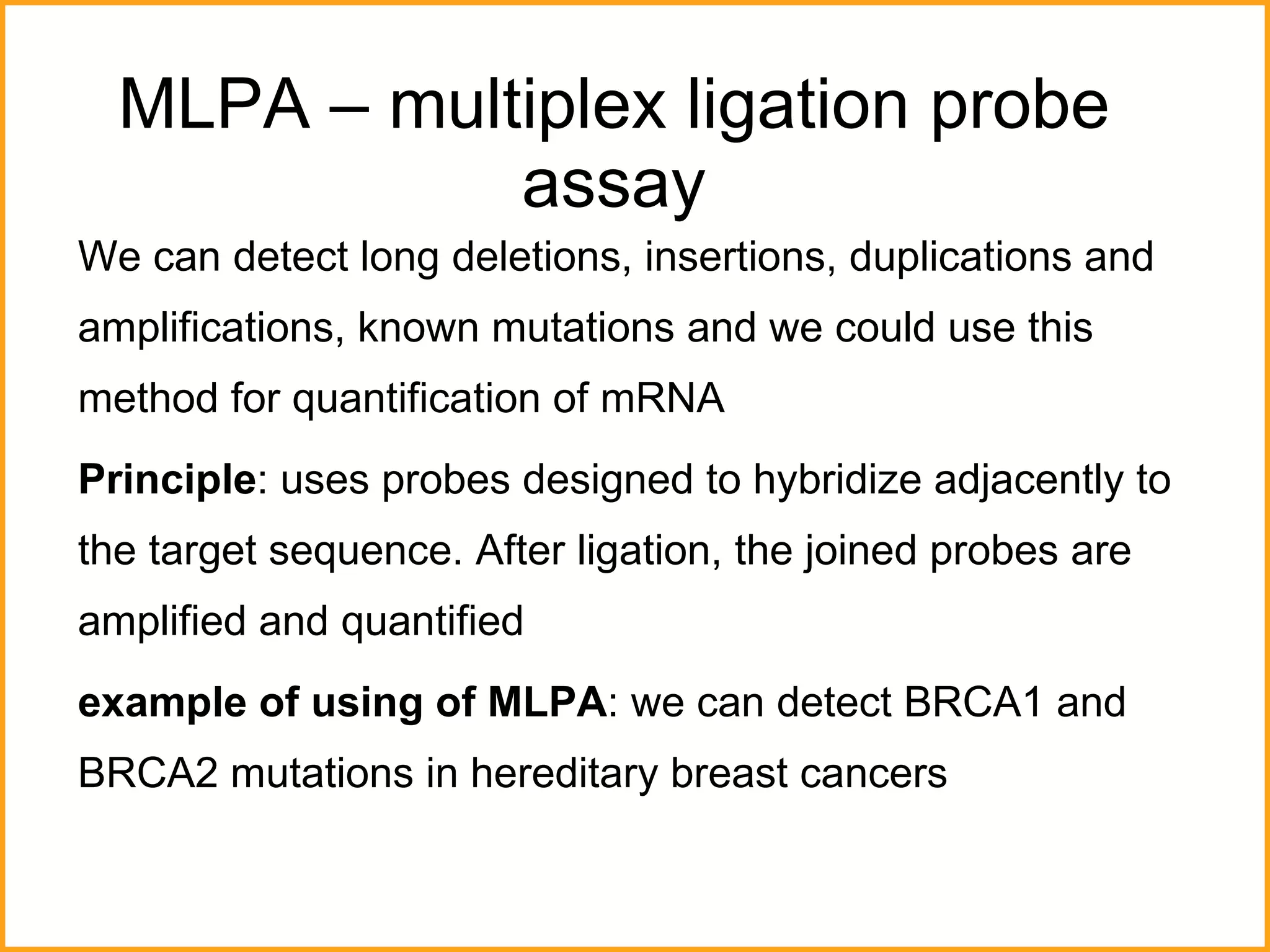
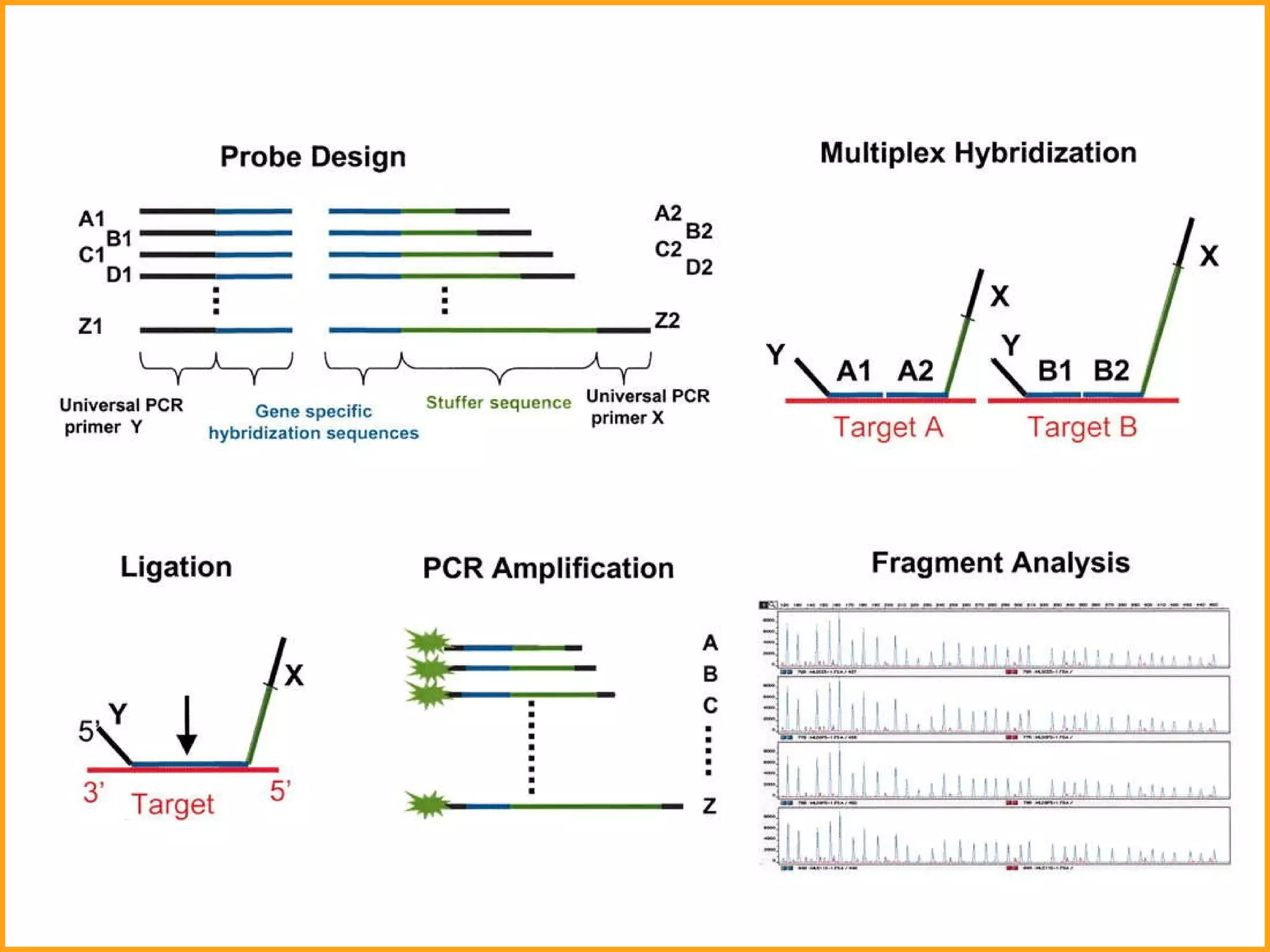
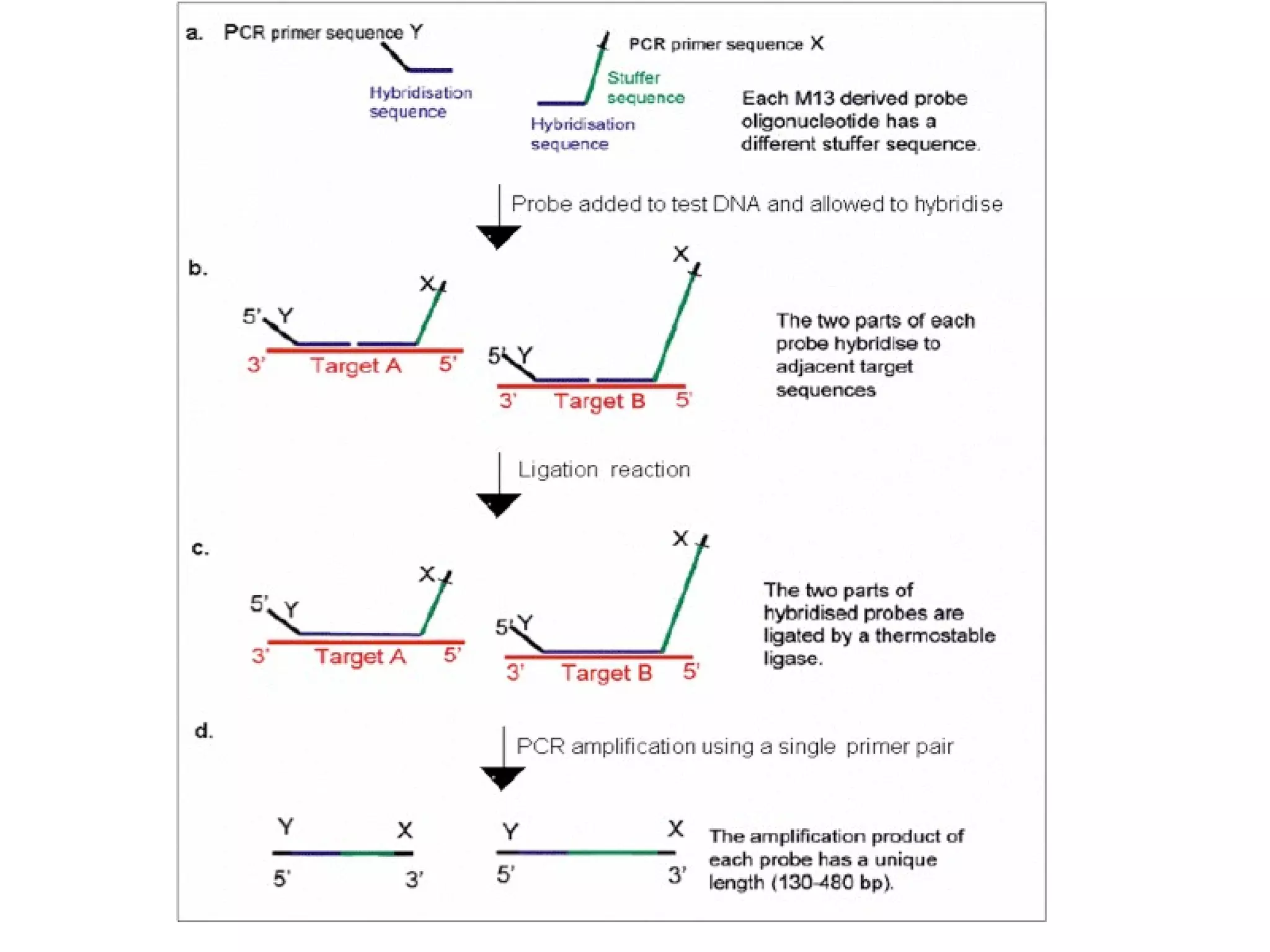
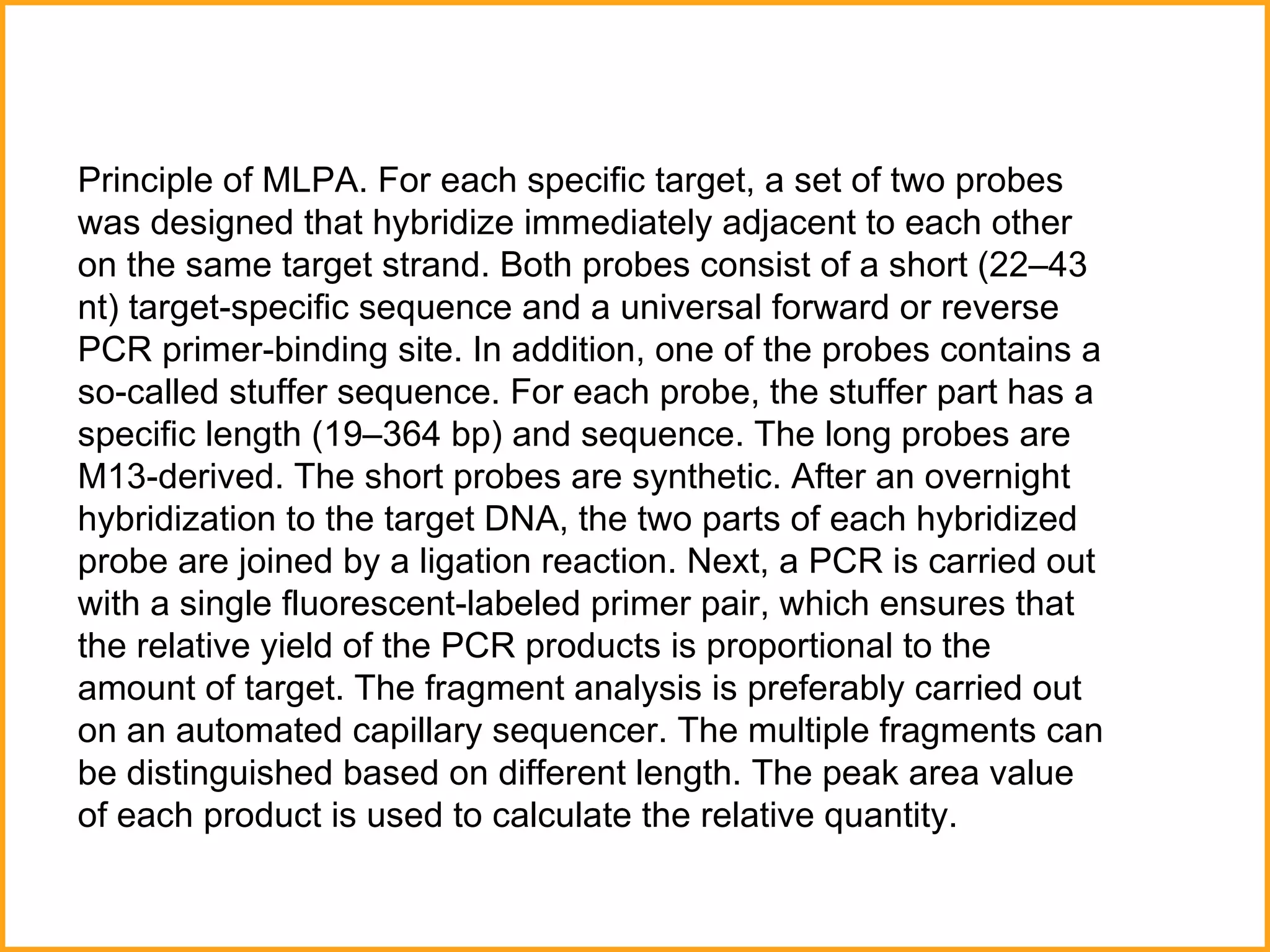
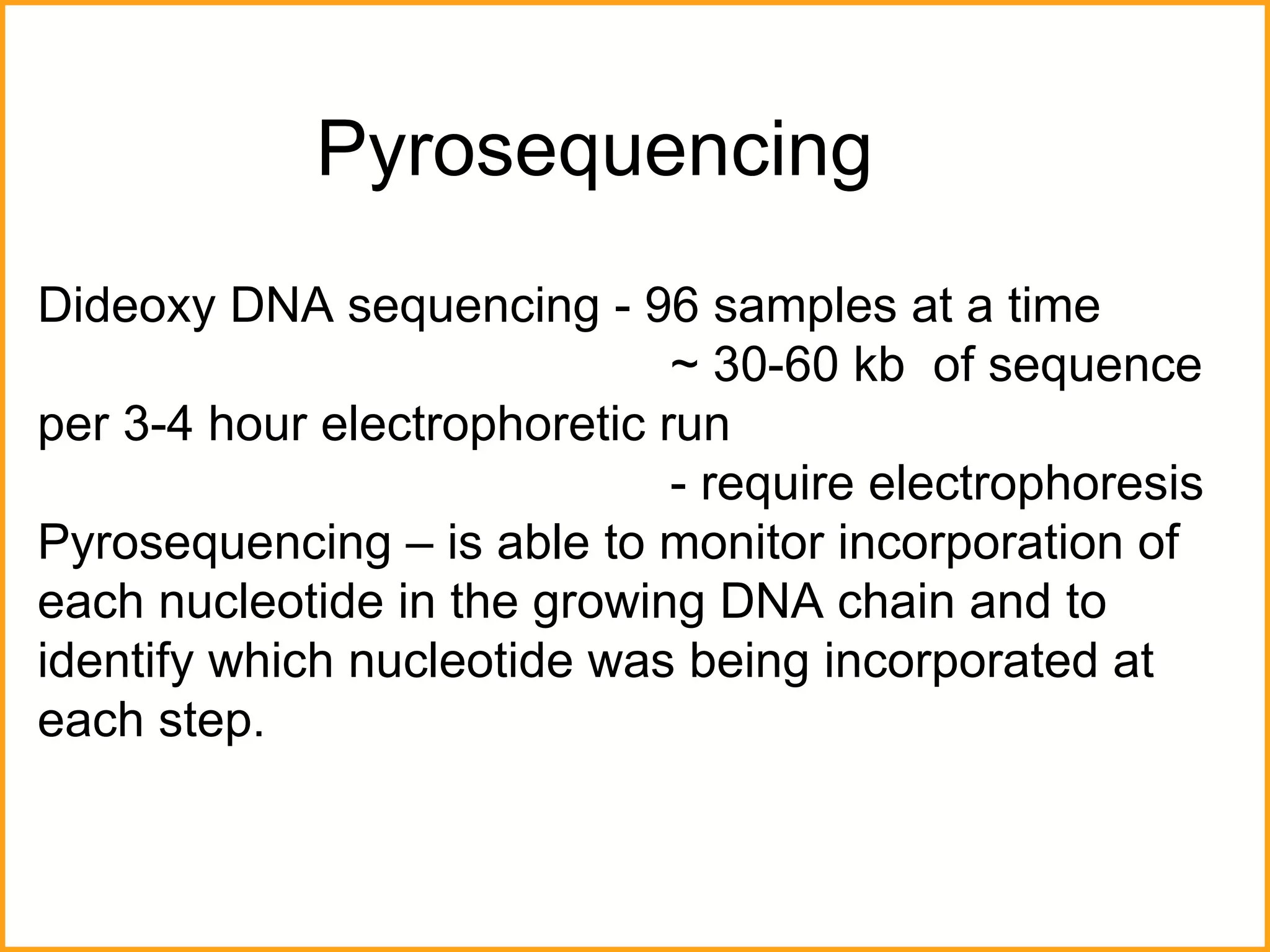
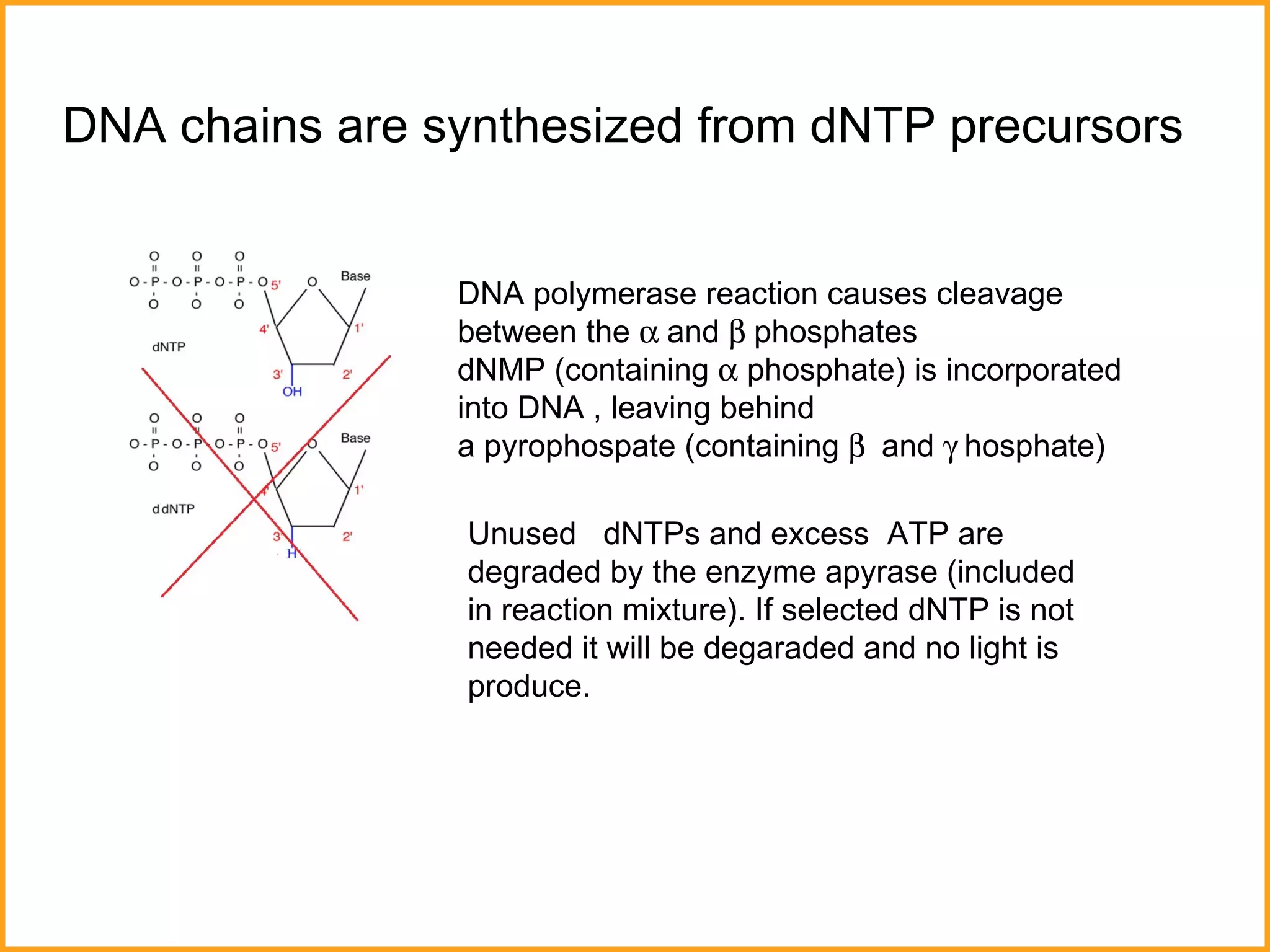
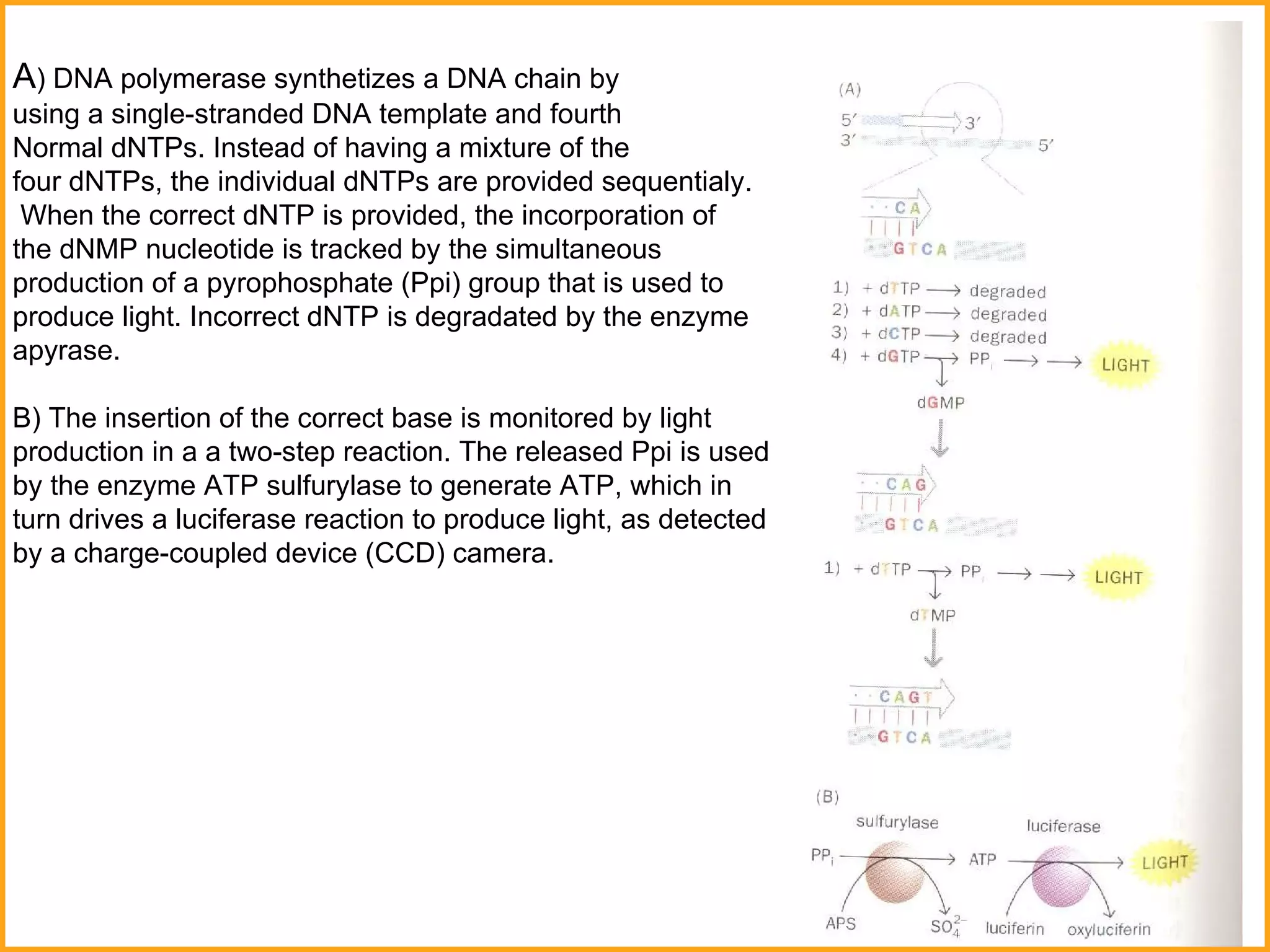
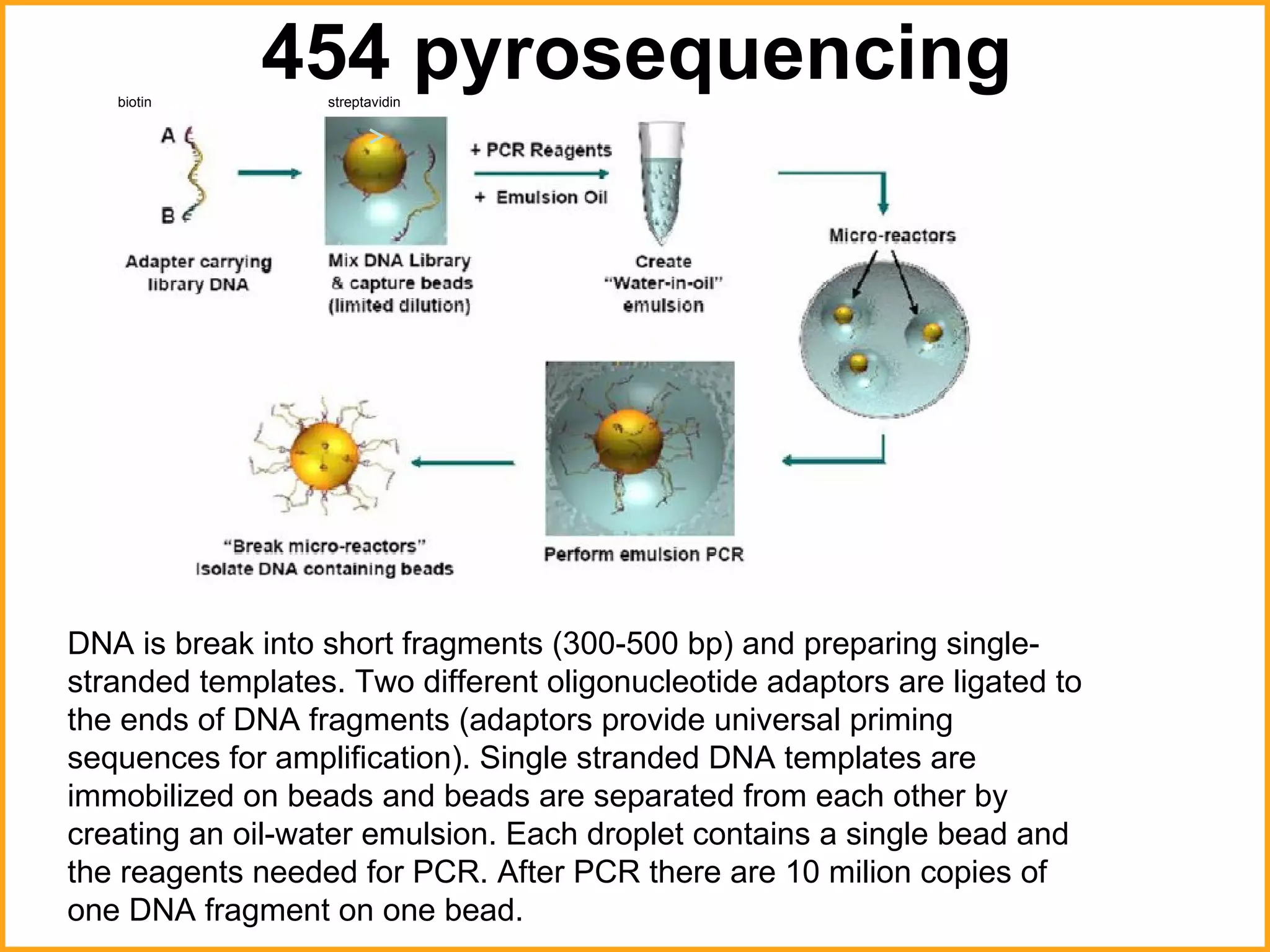
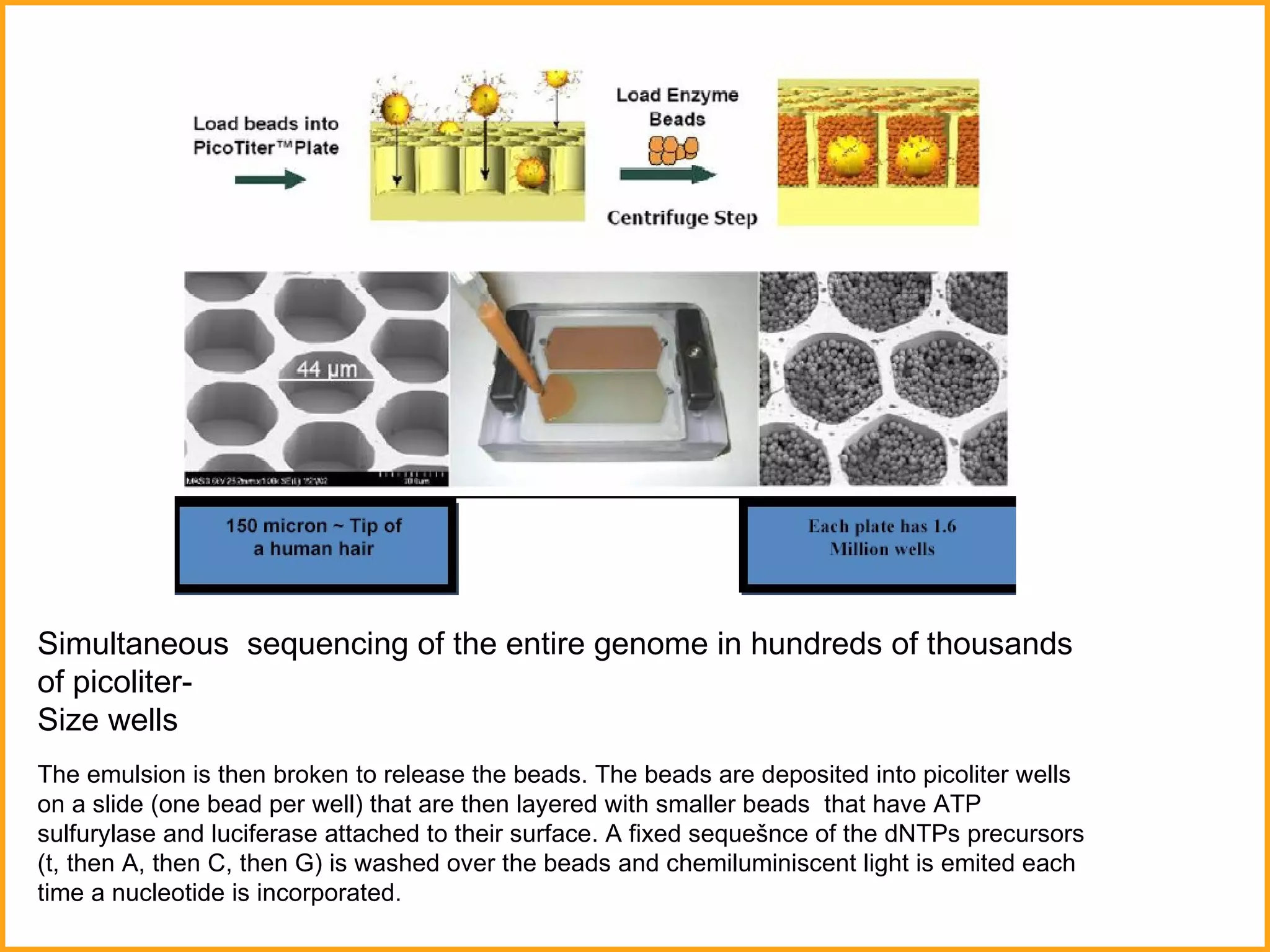
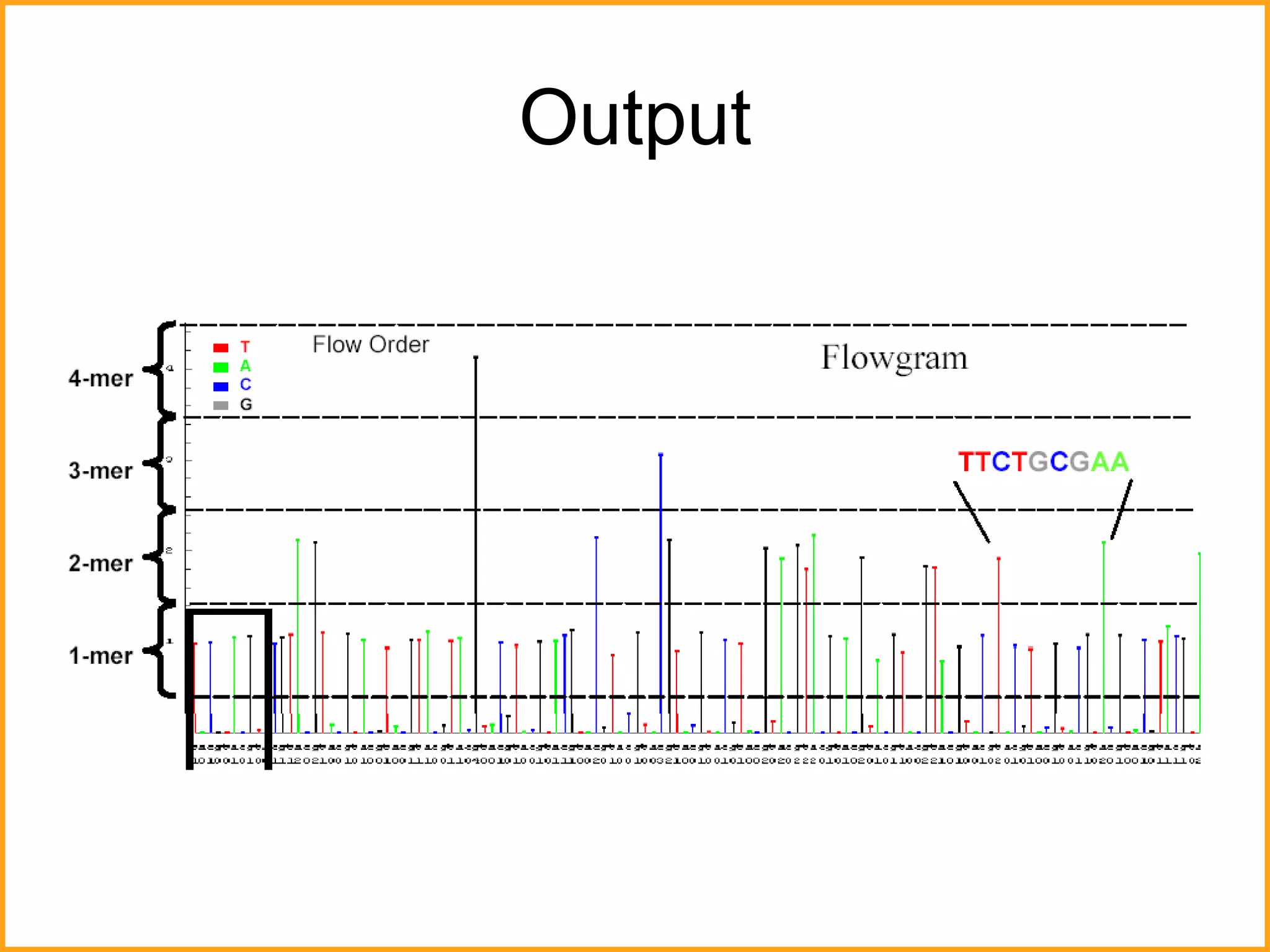
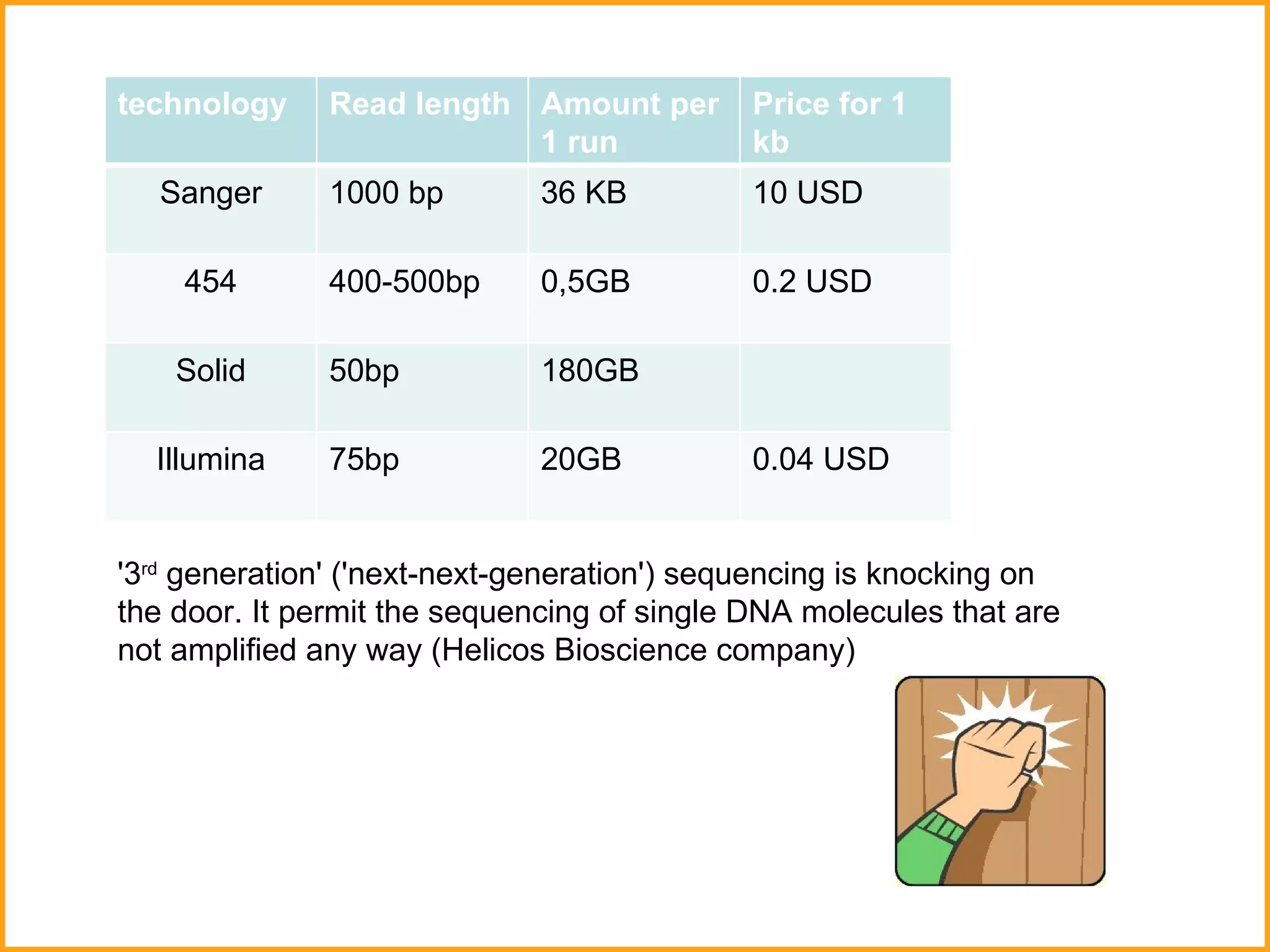
The document discusses various methods in molecular biology, including nucleic acid hybridization, DNA sequencing, real-time PCR, and DNA microarrays. Nucleic acid hybridization uses complementary base pairing between DNA or RNA probes and targets. DNA sequencing determines the nucleotide order using chain-terminating dideoxynucleotides. Real-time PCR quantifies DNA or RNA targets in real time using fluorescent probes. DNA microarrays allow analysis of gene expression patterns across thousands of genes.