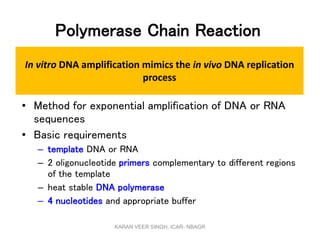
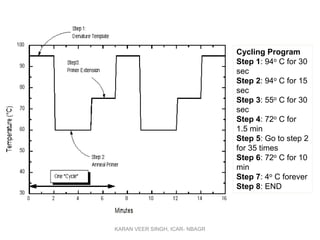
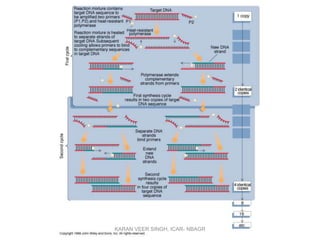
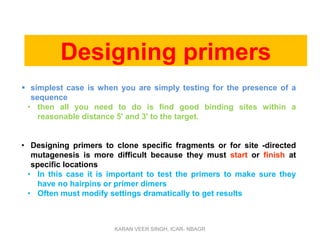
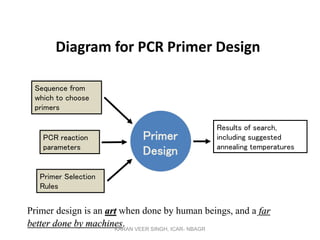
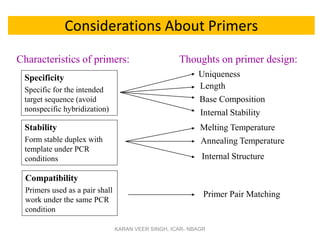
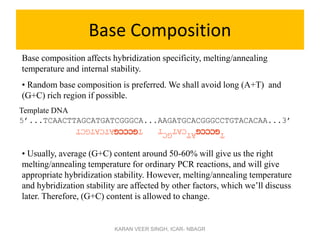
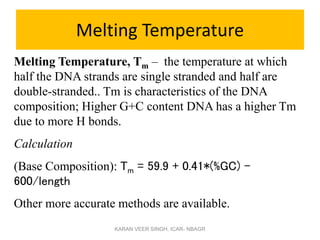
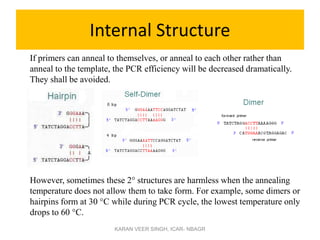
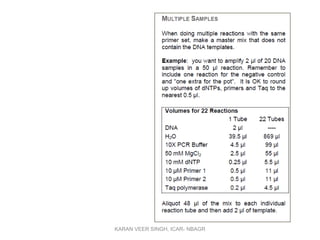
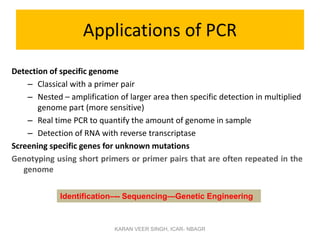
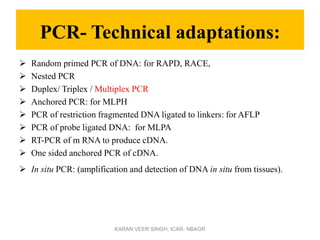
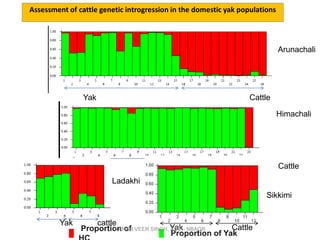
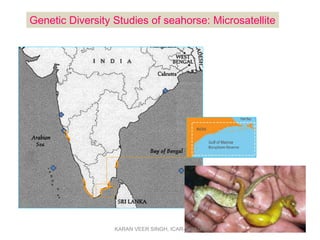
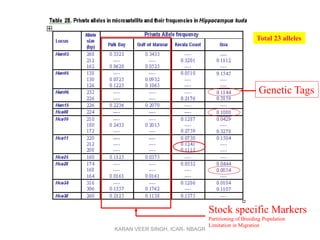
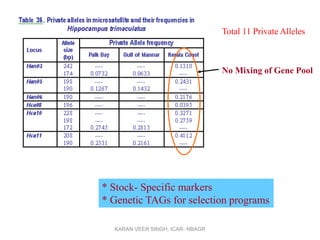
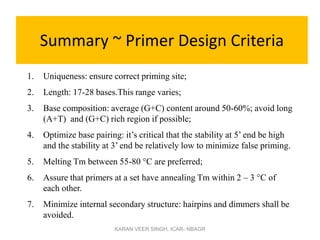
Primer design is a key step in PCR that requires considering various factors to optimize the reaction. These include primer length, melting temperature, GC content, specificity, and potential for secondary structures. Well-designed primers are unique in targeting a single region, have compatible melting temperatures, and do not form hairpins or primer dimers that could inhibit the reaction. Choosing appropriate primers is essential for successful PCR amplification.