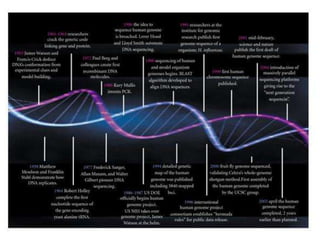
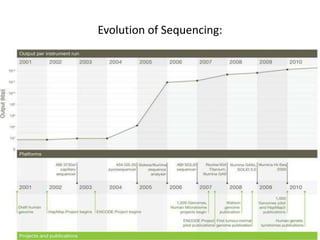
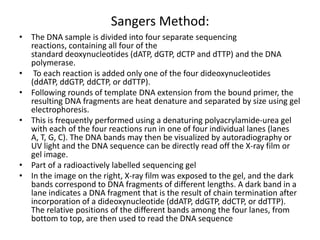
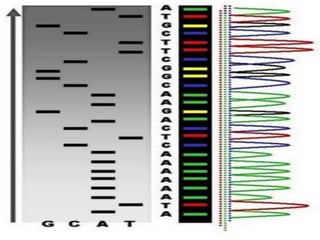
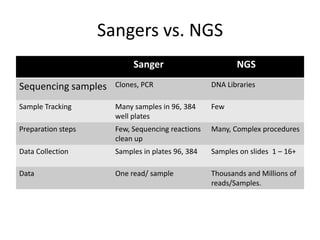
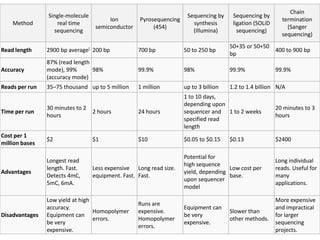
This document provides an overview of next generation sequencing (NGS) technologies. It discusses the history and evolution of DNA sequencing, from early manual methods developed by Sanger to modern high-throughput NGS approaches. Key NGS methods described include Illumina sequencing by synthesis, Ion Torrent semiconductor sequencing, 454 pyrosequencing, and SOLiD ligation sequencing. Compared to Sanger, NGS allows massively parallel sequencing of many samples at lower cost and higher throughput. While NGS has advanced biological research, each method still has advantages and limitations related to read length, accuracy, and cost.