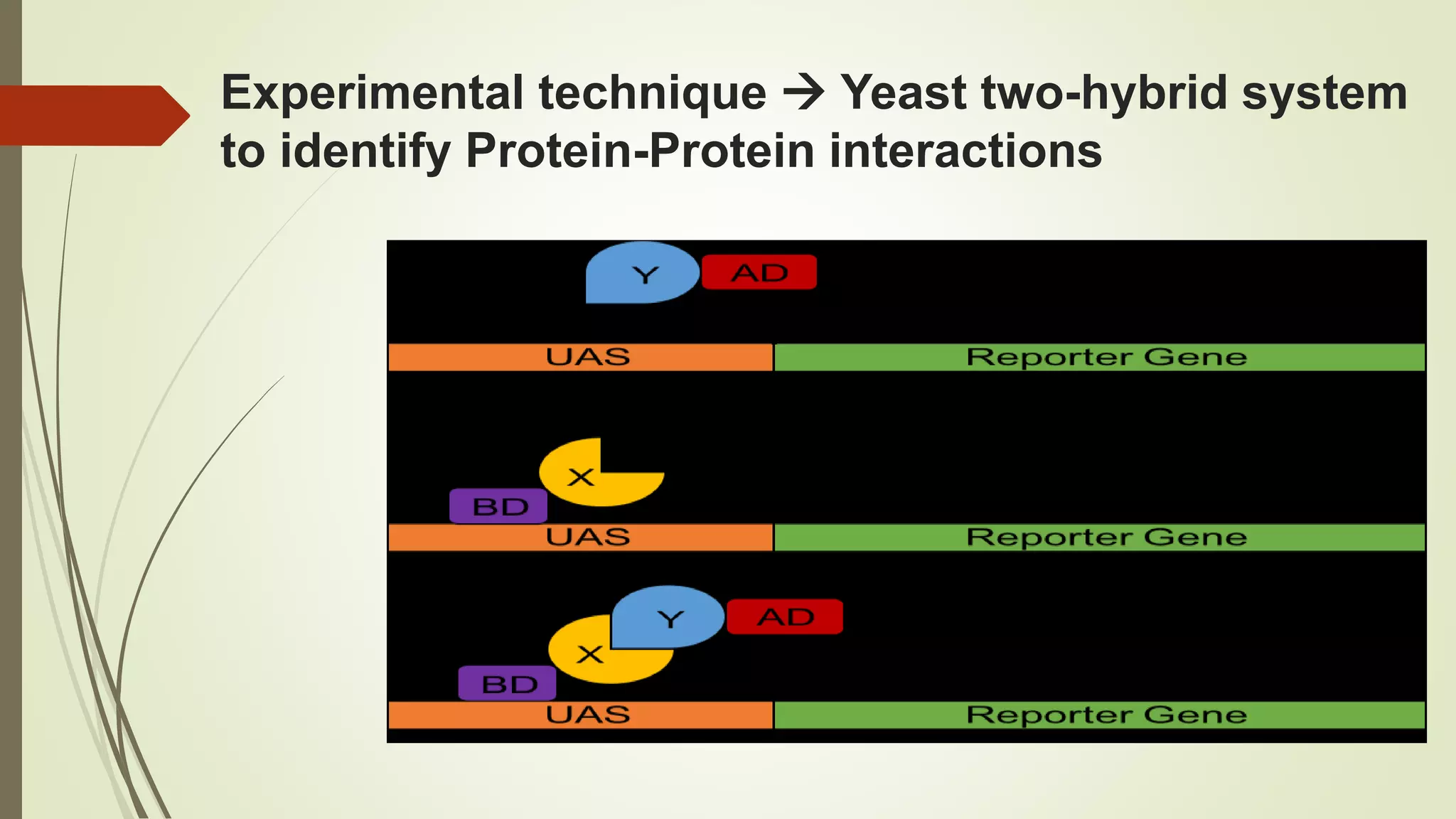
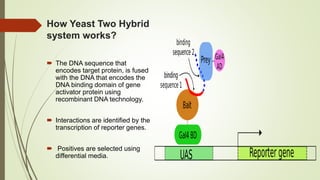
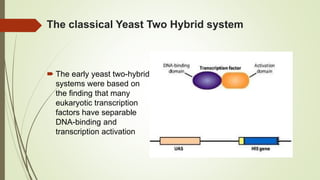
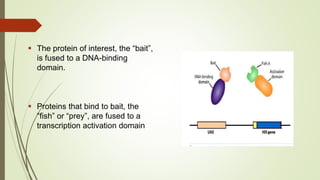
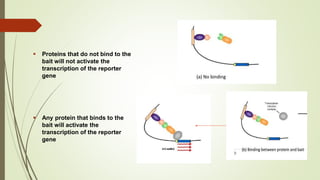
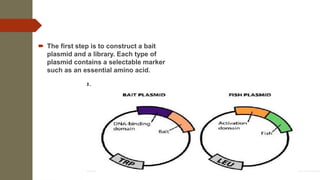
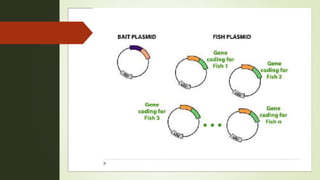
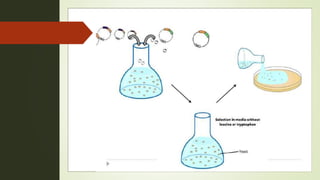
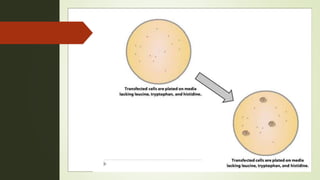
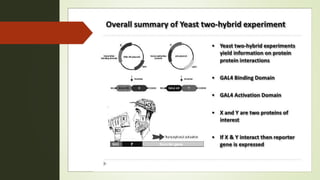
The document discusses the yeast two-hybrid (Y2H) system, a molecular biology technique used to identify protein-protein interactions (PPIs) critical for understanding biological activities and diseases. The Y2H system allows for the detection of these interactions in vivo using reporter genes, providing advantages such as minimal requirements for screening and the ability to detect weak interactions. However, challenges like false positives and potential incorrect protein folding in yeast are also highlighted, along with the conclusion that this method aids in examining cellular processes and discovering pathways.