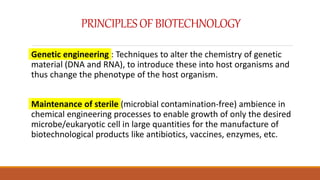
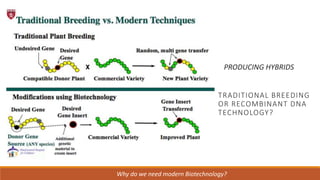
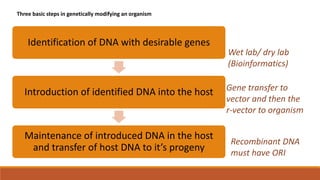
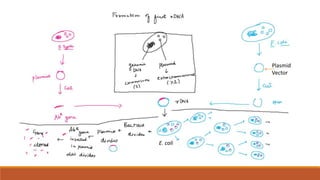
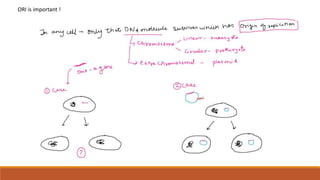
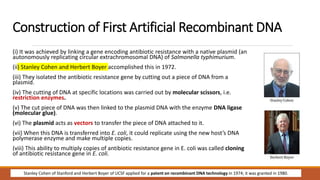
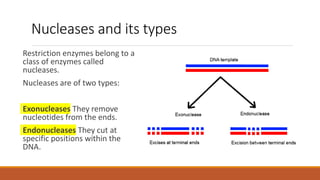
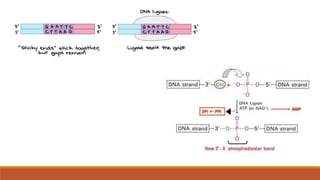
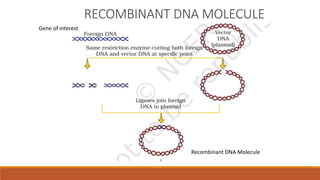
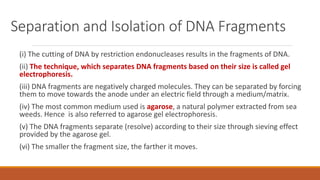
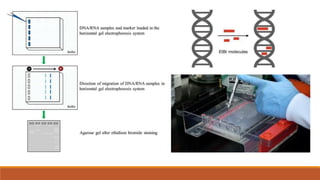
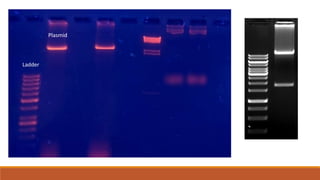
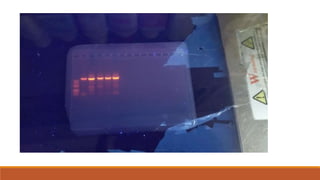
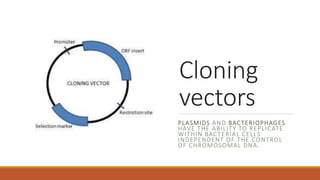
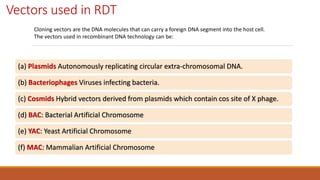
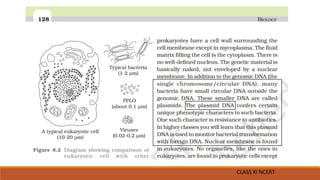
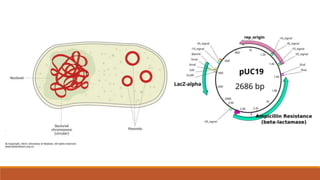
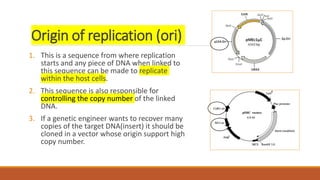
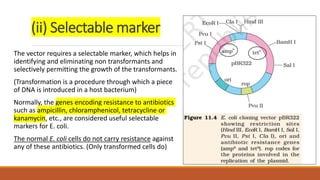
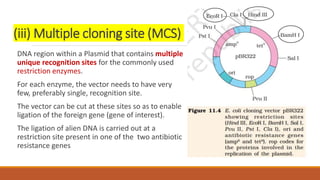
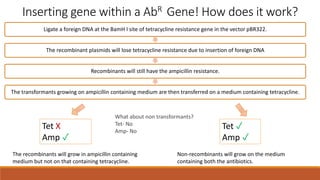
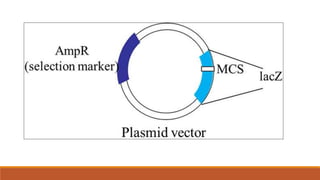
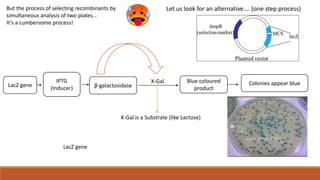
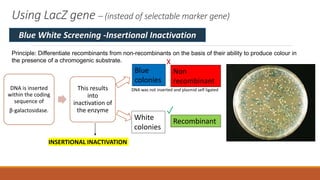
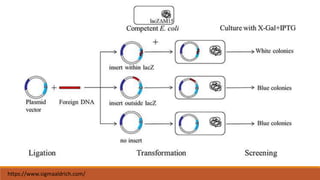
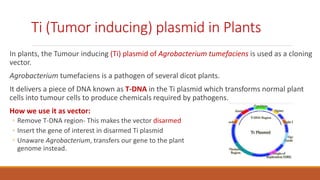
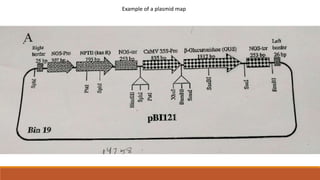
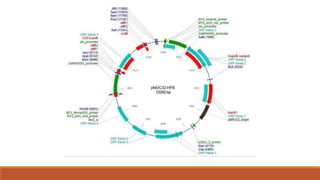
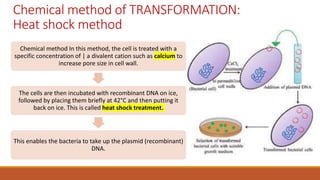
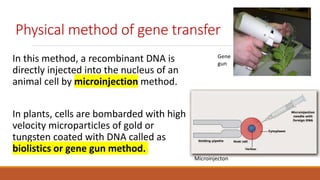
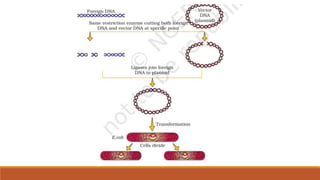
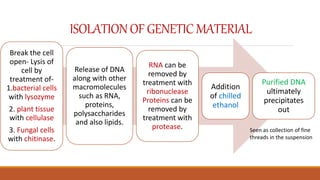
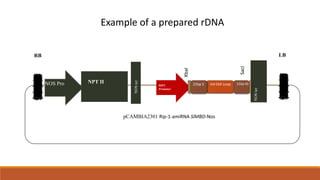
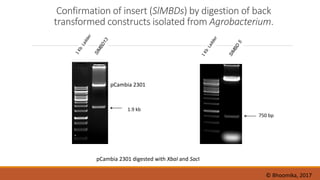
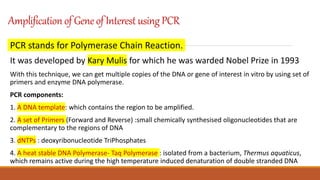
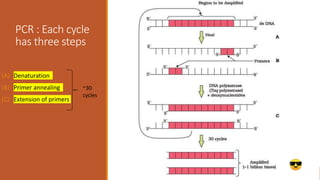
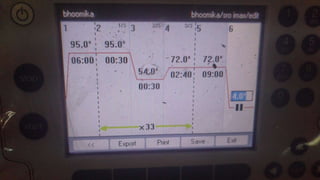
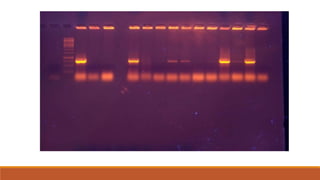
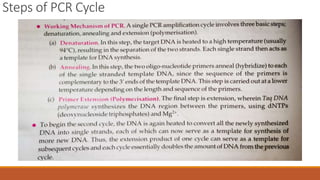
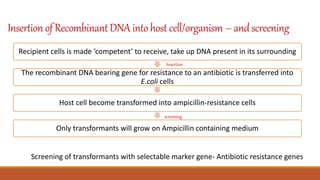
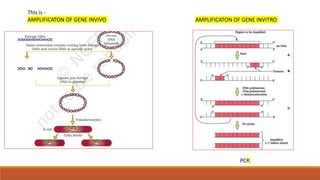
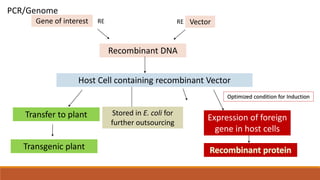
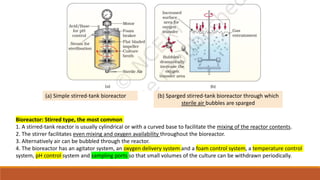
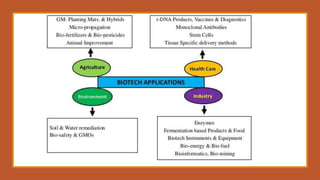
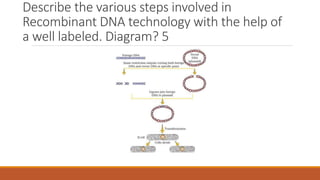
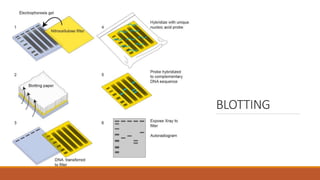
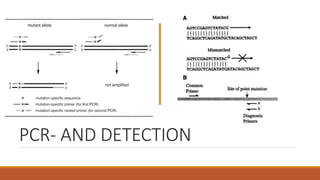
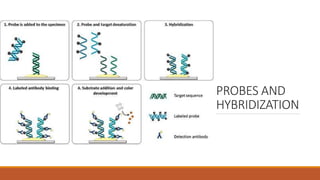
The document elaborates on biotechnology, emphasizing the use of microorganisms, cells, and genetic engineering tools for producing products beneficial to humans. It discusses key concepts and processes in recombinant DNA technology, including the roles of restriction enzymes, cloning vectors, and various techniques for gene transfer and transformation. Additionally, it highlights the differences between traditional and modern biotechnology, focusing on innovations such as artificial recombinant DNA and the use of specific enzymes for genetic manipulation.